FIGURE 4. Short- and long-term replication of HPV16 subgenomic DNA in the pGAB1 vector.
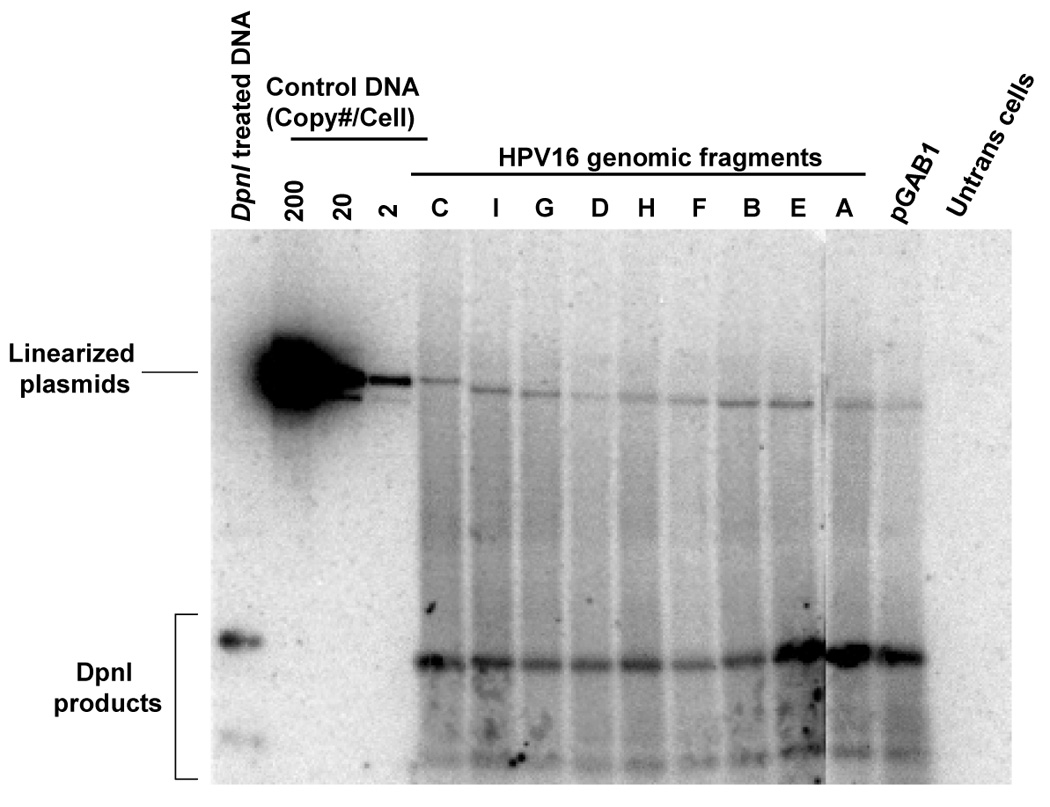
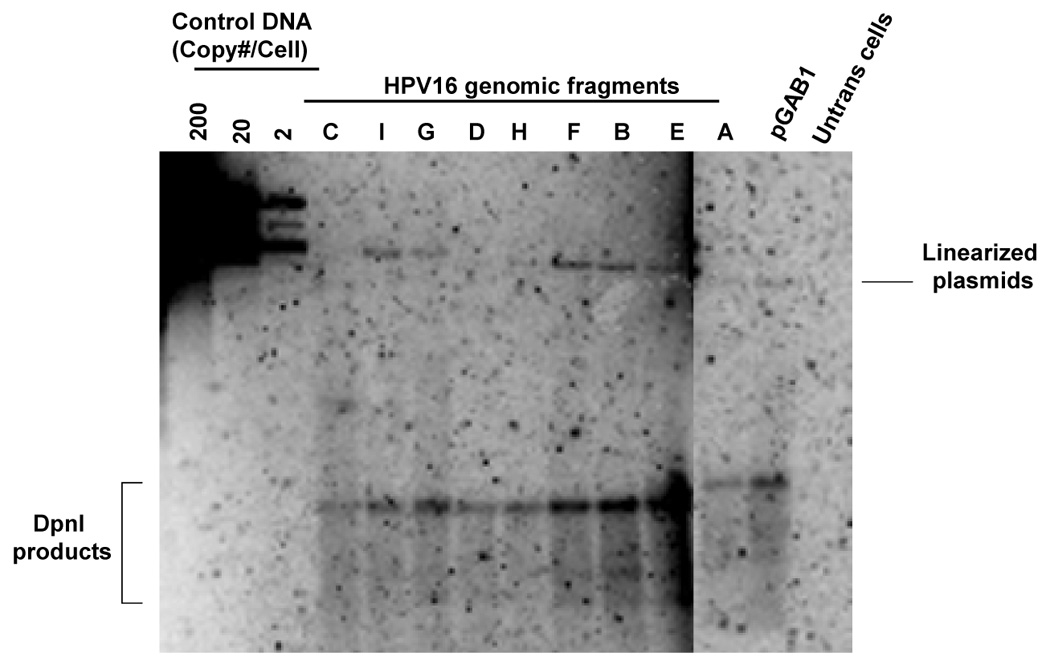
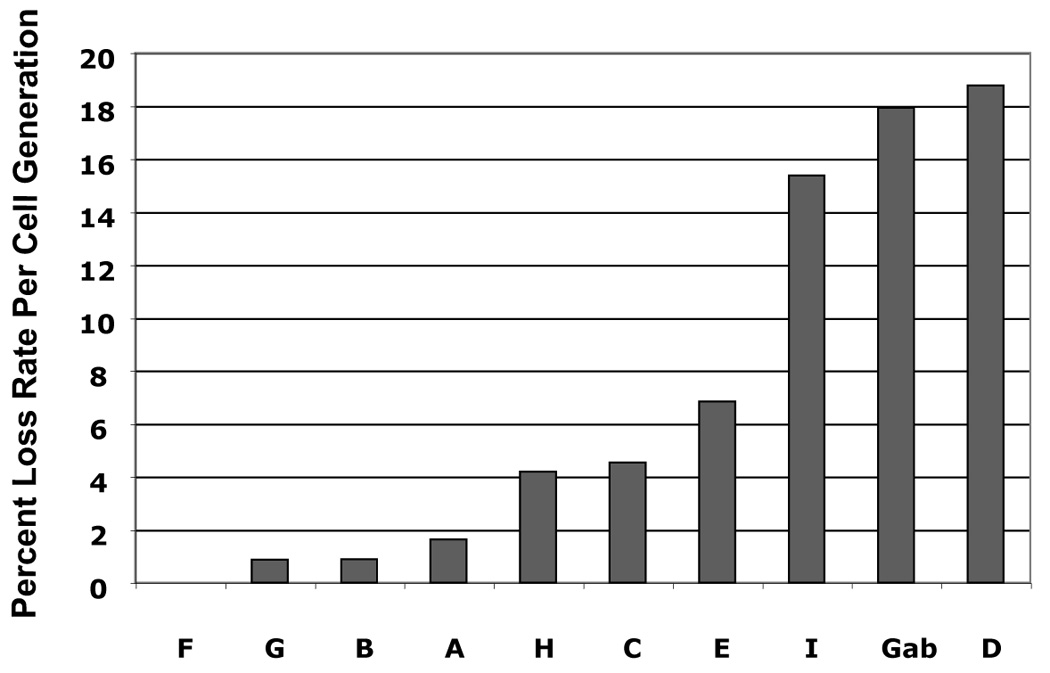
(A) DpnI-resistant newly synthesized DNAs were linearized with XhoI and detected by Southern blot at 4 days posttransfection. XhoI–treated DNAs indicated copy number were electrophoresed alongside. At the far left, DpnI-treated control DNA showed the completeness of DpnI cutting of DpnI-sensitive DNA from bacteria. (B) Southern analysis of Hirt extracted DNAs at 11 days after transfection. (C) The stability of plasmids containing HPV16 subgenomic fragments was demonstrated as the relative loss rate per cell generation. The percent of GFP-positive cells was determined by flow cytometry at different time points. The change in percent GFP-positive cells per cell generation (loss rate) was computed relative to the first time point of each transfection. The loss rates of each plasmid were normalized with that of the most stable plasmid to obtain the relative loss rate per cell generation.
