Abstract
Purpose:
Somatic alterations have been shown to correlate with ovarian cancer prognosis and survival, but less is known about the effects on survival of common inherited genetic variation. Of particular interest are genes involved in cell cycle pathways, which regulate cell division and could plausibly influence clinical characteristics of multiple tumors types.
Experimental Design:
We examined associations between common germ-line genetic variation in 14 genes involved in cell cycle pathway (CCND1, CCND2, CCND3, CCNE1, CDKN1A, CDKN1B, CDKN2A, CDKN2B, CDKN2C, CDKN2D, CDK2, CDK4, CDK6, and RB1) and survival among women with invasive ovarian cancer participating in a multicenter case-control study from United Kingdom, Denmark, and United States. DNAs from up to 1,499 women were genotyped for 97 single-nucleotide polymorphisms that tagged the known common variants (minor allele frequency ≥0.05) in these genes. The genotypes of each polymorphism were tested for association with survival by Cox regression analysis.
Results:
A nominally statistically significant association between genotype and ovarian cancer survival was observed for polymorphisms in CCND2 and CCNE1. The per-allele hazard ratios (95% confidence intervals) were 1.16 (1.03-1.31; P = 0.02) for rs3217933, 1.14 (1.02-1.27; P = 0.024) for rs3217901, and 0.85 (0.73-1.00; P = 0.043) for rs3217862 in CCND2 and 1.39 (1.04-1.85; P = 0.033) for rs3218038 in CCNE1. However, these were not significant after adjusting for multiple hypothesis tests.
Conclusion:
It is unlikely that common variants in cell cycle pathways examined above associated with moderate effect in survival after diagnosis of ovarian cancer. Much larger studies will be needed to exclude common variants with small effects.
Ovarian cancer is the seventh most common malignancy occurring in women and causes 125,000 deaths annually in women worldwide (1). It is the leading cause of death from gynecologic malignancy accounting for 4% of new cancer cases and 4.3% of all female deaths from cancer. Ovarian cancer is often advanced at presentation and is associated with a poor prognosis. Despite advances in treatment of ovarian cancer over the past 20 years, the overall 5-year survival is still <30%.
Somatic alterations and rare germ-line mutations in high-penetrance susceptibility genes have been shown to correlate with ovarian cancer prognosis and survival (2, 3), but the effects of common inherited genetic variation are less well understood. Single-nucleotide polymorphisms (SNP) may have clinic pathologic importance as prognostic markers. If such germ-line genetic markers of prognosis can be reliably identified, they have the potential to improve on the prediction of outcome for ovarian cancer as well as offer insights into the biological determinants of response to treatment and prognosis.
Several studies investigating common genetic variation and prognosis in ovarian cancer have been published (4-17), some of which have reported significant associations at a nominal P < 0.05 (7-14, 16). However, most of these studies were small (<220 cases), and none reached the level of statistical significance that has been suggested as appropriate for genetic association studies in candidate genes (P < 10−4; ref. 18).
Cell cycle checkpoints are part of the regulatory pathways that control the order and timing of cell cycle transitions to ensure the fidelity of critical events such as DNA replication and chromosome segregation (19). Disruption of cell cycle control pathways has previously been related to the prognosis of human cancers (20). For example, CCND1 variants were associated with prognosis in non–small-cell lung cancer, breast cancer, and squamous cell carcinoma of the head and neck, but were not associated with overall prognosis of epithelial ovarian cancer (21-24). In addition, CDKN2A and RB1 expression levels have been associated with prognosis for advanced-stage ovarian cancer (25).
We have previously investigated the association of 97 tag SNPs that capture the common variation in 14 genes involved in G1-S phase progression (CCND1, CCND2, CCND3, CCNE1, CDK2, CDK4, CDK6, CDKN1A, CDKN1B, CDKN2A, CDKN2B, CDKN2C, CDKN2D, and RB1) with susceptibility to invasive epithelial ovarian cancer (26, 27). We found four border-line significant associations with ovarian cancer susceptibility: RB1 rs2854344 (P = 0.0015), RB1 rs4151620 (P = 0.0001), CDKN2A rs3731257 (Ptrend = 0.008), and CDKN1B rs2066827 (Ptrend = 0.036). The purpose of the current study was to test the hypothesis that germ-line genetic variation in cell cycle control genes affects survival after diagnosis of ovarian cancer. To do this, we have linked the genetic data from the analysis of the 14 cell cycle control genes described above to outcome data from regional cancer registries in ∼1,500 invasive ovarian cancer cases.
Materials and Methods
Study population
The study subjects were from three different ovarian cancer case-control studies: the Studies of Epidemiology and Risk Factors in Cancer Heredity (SEARCH) study from the United Kingdom, the Malignant Ovarian Cancer (MALOVA) study from Denmark, and the Family Registry for Ovarian Cancer (FROC) study from the United States (Table 1).
Table 1.
Characteristics of ovarian cancer cases
| Total | SEARCH | FROC | MALOVA | |
|---|---|---|---|---|
| Total no. subjects | 1,499 | 726 | 327 | 446 |
| Total time at risk, person-years | 6,013 | 2,998 | 1,210 | 1,804 |
| Median follow-up, y* | 6.15 (0.01-10)† | 7.61 (1.89-10) | 4.54 (0.27-8.27) | 3.27 (0.01-9.72) |
| Median time at risk, y | 4.16 (0.00-9.72)† | 4.69 (0.10-6.58) | 3.67 (0.00-7.74) | 3.26 (0.01-9.72) |
| Median time from diagnosis to study entry, y | 1.39 (0.00-8.77)† | 2.7 (0.34-8.77) | 0.55 (0.15-4.04) | 0.00 (0.00-0.32) |
| No. deaths | 637 | 189 | 147 | 301 |
| Annual mortality rate | 0.11 | 0.063 | 0.12 | 0.17 |
| Median 5-y survival (95% CI), % | 48 (44-51)‡ | 72 (68-77) | 52 (45-58) | 40 (35-44) |
| Median age at diagnosis, y | 56 (21-80)† | 56 (21-74) | 51 (23-64) | 60 (32-80) |
| Age at diagnosis (y), n (%) | ||||
| <40 | 102 (7) | 50 (7) | 39 (12) | 13 (3) |
| 40-49 | 297 (20) | 130 (18) | 96 (29) | 71 (16) |
| 50-59 | 529 (35) | 273 (38) | 126 (39) | 130 (29) |
| >60 | 571 (38) | 273 (38) | 66 (20) | 232 (52) |
| Total | 1,499 (100) | 726 | 327 | 446 |
| Histopathologic type,§ n (%) | ||||
| Serous | 698 (51) | 257 (40) | 166 (58) | 275 (62) |
| Endometrioid | 234 (17) | 131 (20) | 47 (16) | 56 (13) |
| Mucinous | 169 (12) | 97 (15) | 29 (10) | 43 (10) |
| Clear cell | 118 (9) | 62 (10) | 23 (8) | 33 (7) |
| Papillary NOS | 77 (6) | 38 (6) | 9 (3) | 30 (7) |
| Other | 78 (6) | 57 (9) | 13 (5) | 8 (2) |
| Total known | 1,374 | 726 | 327 | 445 |
| Unspecified | 125 | 84 | 40 | 1 |
| Clinical stage, n (%) | ||||
| Localized tumor | 561 (37) | 291 (40) | 122 (37) | 148 (33) |
| Advanced disease∥ | 629 (42) | 143 (20) | 188 (57) | 298 (67) |
| Total known | 1,190 | 434 | 310 | 446 |
| Unknown | 309 | 292 | 17 | 0 |
| Grade, n (%) | ||||
| Well differentiated | 251 (17) | 102 (14) | 45 (14) | 104 (23) |
| Moderately differentiated | 376 (25) | 167 (23) | 64 (20) | 145 (33) |
| Poorly/undifferentiated | 504 (34) | 182 (25) | 154 (47) | 168 (38) |
| Total known | 1,131 | 451 | 263 | 417 |
| Unknown | 368 | 275 | 64 | 29 |
Follow-up censored at 10 y.
Range of variable.
95% CI.
The calculation of percentage of histopathologic subtype was using the number of total known as denominator in each column respectively.
Spread to regional lymph nodes or distant metastases.
The SEARCH ovarian cancer study is an ongoing, population-based ovarian cancer case control study covering the regions served by the East Anglia and West Midlands cancer registries in the United Kingdom. All patients diagnosed in East Anglia with invasive epithelial ovarian cancer since 1991, of ages <70 years, and still alive in 1998 when recruitment started are invited to take part (prevalent cases). Incident cases are those diagnosed since 1998 in East Anglia and since 2003 in the West Midlands, of ages <70 years. From 1991 to the end of 1997, 1,181 women were registered ovarian cancer cases in East Anglia, of whom 767 had already died; the general practitioners refused permission to contact 166 patients. Thus, we invited 248 women to take part, of whom 216 provided a blood sample (87% of those invited and 18% of all eligible diagnoses in the region). As the study is ongoing, the following data are based on registrations in East Anglia from 1998 to 2004 for which recruitment has been completed. In this period, 1,453 women were registered, of whom 334 had died by the time of registration and the general practitioner refused permission to contact 531 (reasons unknown). Of 588 women invited to take part, 476 provided a blood sample (81% of those invited and 33% of all eligible diagnoses). To date, we have invited 1,750 women to participate, of whom 1,157 have provided a blood sample—the first 726 cases were available for this analysis. The study is approved by the Eastern Multicenter Research Ethics Committee. DNA was extracted from blood samples by Whatman International Ltd.
The MALOVA study is a population-based, Danish case-control study of ovarian cancer. Eligible cases were women ages 30 to 80 years, who were diagnosed with an ovarian tumor from December 1994 to May 1999. The study included 18 different hospitals from the municipalities of Copenhagen and Frederiksberg as well as the counties of Copenhagen, Frederiksborg, Roskilde, Western Sealand, Storstrøm, Funen, Southern Jutland, and Northern Jutland. By the end of the study period, a total of 959 ovarian cancer patients were identified in this study area. Of these, 53 patients were considered too ill to participate; 45 women died before being contacted. Thus, 861 were invited to take part, of whom 652 (76%) provided a blood sample. Samples were collected at the time of diagnosis. Samples from 446 invasive cases were available for this study. DNA was extracted from blood samples by Whatman International. This study has been approved by the scientific ethical committee in the study area (KF01-384/95) and all subjects provided written consent.
The U.S. subjects were ascertained from six counties in Northern California as part of the FROC study. Patients with epithelial ovarian cancer diagnosed between March 1, 1997 and July 31, 2001 were identified through the Greater Bay Area Cancer Registry operated by the Northern California Cancer Center as part of the Surveillance, Epidemiology, and End Results Program of the National Cancer Institute. Rapid case ascertainment was used to identify cases within 1 month of diagnosis. Eligible patients were those diagnosed with invasive or low malignant potential epithelial ovarian cancer, ages 20 to 64 years, who resided in any of six Bay Area counties (Alameda, Contra Costa, Marin, San Francisco, San Mateo, or Santa Clara). Of the 915 eligible patients, 24 were not contacted because their physicians refused permission; 209 refused to participate, had died, or could not be located; and 682 (75%) were interviewed. Of these, 579 (85% of those interviewed and 63% of total) genomic DNAs were isolated from leucocytes of peripheral blood using the Puregene Kit (Gentra Systems). Genomic DNA was also isolated from exfoliated cells in buccal mouthwash rinses as previously described (28). Research was conducted with protocols approved by the Institutional Review Boards of Stanford University School of Medicine and Roswell Park Cancer Institute. This analysis is restricted to the 327 White cases with invasive epithelial ovarian cancer, for whom DNA was available.
Follow-up
Active follow-up was carried out for SEARCH participants by the Eastern and West Midland cancer registries at 3 and 5 years after diagnosis, and then at 5-year intervals. This is done by searching hospital information systems for recent visits. When it is observed that a patient has had no recent visit, the general practitioner of the patient is contacted to ascertain the patient's vital status. In addition, the registries obtain notification of deaths through death certificate flagging with the Office for National Statistics. There is a lag time with this process of a few weeks for cancer deaths and 2 months to a year for noncancer deaths. At the time of analysis, 189 patients had died within 10 years of diagnosis.
FROC did active follow-up of subjects until 2002. Updates of the vital status from the Greater Bay Area Cancer Registry, which is operated by the Northern California Cancer Center as part of the National Cancer Institute Surveillance, Epidemiology, and End Results Program, were carried out for patients once or twice during the study and most recently in 2004. In the state of California, cancer is a state-mandated reportable condition, and the cancer registry for the greater Bay Area has been in existence since 1973. It is from this group that all investigators who want to do population-based cancer studies in this area receive their data. Staff periodically review computerized hospital tumor registry data or medical records for updated information. They also receive updates on the person's vital status from the state's death index (which is usually 18 months behind for reporting).
MALOVA did active follow-up until 2003. In Denmark, all inhabitants have a unique personal (10-digit) identification number (CPR number), which is used universally in the Danish society. These identification numbers, which comprise information on date of birth and sex, are registered in the computerized Danish National Central Population Register. The register contains information on, for example, dates of death and emigration. All cases in this study were traced in the register and “date of death,” “emigration,” or “May 2003,” whichever came first, was registered. Hospital files were collected from all patients in the study. Women who died during follow-up were linked to a Danish Hospital Reference System and information about exact cause of death was assessed in each case by matching information in this register with the clinical records.
Selection of genes and SNPs
The 14 genes were selected as candidate genes for ovarian cancer susceptibility. We used a comprehensive SNP tagging approach in which tag SNPs were chosen to capture all the known common genetic variation in each gene with a minimum correlation coefficient (r2) of 0.8. Data from the International Hap Map project9 and resequencing data from the Environmental Genome Project10 were used to select tag SNPs. In total, 97 SNPs were chosen to tag 224 common variants [see Song et al. (26) and Gayther et al. (27) for details].
Genotyping
A detailed description of the genotyping has already been published for these genes (26, 27). Briefly, all of samples were genotyped using the TaqMan 7900HT Sequence Detection System according to manufacturer's instructions. Primer and probe sequences and assay conditions used for each polymorphism analyzed are available on request. All assays were carried out in 384-well plates and included 12 duplicate samples in each plate for quality control. Genotypes were determined using Allelic Discrimination Sequence Detection Software (Applied Biosystems). Genotyping of SEARCH and FROC samples was carried out at the Department of Oncology, University of Cambridge, and MALOVA samples were genotyped at the Translational Research Laboratory, University of College London. Call rates ranged from 91% to 99.8% for all the individual studies for all the SNPs (except rs4151620, rs2854344, and rs3092904, which failed in partial plates of MALOVA study) and overall concordance between duplicate samples was >98%. Individual samples with failed calls were not repeated. Hence, there are variations in the number of samples successfully genotyped for each polymorphism.
Statistical methods
The effect of each SNP on survival was assessed using Cox regression analysis stratified by study. Because samples were taken at a variable time after diagnosis for SEARCH and FROC studies, analyses were conducted allowing for left-truncated data. Time at risk began at the date of blood sample receipt and ended at the date of death from any cause, or, if death did not occur, on the last time known to be alive. Follow-up for all cases was censored at 10 years after diagnosis. The primary tests were the likelihood ratio test for trend (1 degree of freedom) based on the number of rare alleles carried. The hazard ratio (HR) per rare allele carried was also estimated from the Cox regression. We also investigated possible effects of combination of alleles that tagged specific SNPs (multimarker haplotypes). For specific haplotype marker tests, haplotype frequencies and subject-specific expected haplotype indicators were calculated separately for each study using the program TagSNPs (29). This implements an expectation substitution approach to account for haplotype uncertainty given unphased genotype data. Subjects missing >50% genotype data were excluded from haplotype analysis. We used unconditional logistic regression to test the null hypothesis of no association between specific multimarker tagging haplotype and survival by comparing a model with terms for subject-specific haplotype indicator with an intercept-only model. The assumption of proportional hazards was assessed graphically by log-log survival curves.
Data on histopathologic subtype, tumor stage, grade, and age at diagnosis were available for 79%, 99.6%, 75%, and 100% of the cases, respectively (Table 1). This enabled us to evaluate the significance of each polymorphism after adjusting for known prognostic factors. Factors were grouped as follows: age at diagnosis (<40, 40-49, 50-59, and >60 years); tumor stage (localized and advanced); grade (well differentiated, moderately differentiated, and poorly/undifferentiated); and histologic subtype [serous, clear cell, endometrioid, mucinous, papillary NOS (not otherwise specified), and other]. The SNPs significantly associated with survival at the 5% level were retested in multivariate analysis models to adjust for “true” stratification for prognostic factors (as opposed to adjustment by inclusion of dummy covariates), so that baseline hazard could vary by strata. All analyses were done with Stata version 8.0.
Results
Association of genotype with survival
The characteristics of the ovarian cancer study participants from SEARCH, FROC, and MALOVA are described in Table 1. More than 99% of the cases were White. During the 6,013 person-years at risk, there were 637 deaths.
We identified 19 SNPs with a P ≤ 0.2, of which 4 SNPs had a P ≤ 0.05 in test for association. The results of the univariate Cox regression analyses are summarized in Table 2. The complete data for all SNPs are given in Supplementary Table S1. None of the SNPs in RB1, CDK2, CDK4, CDK6, CDKN2A, CDKN2B, CDKN2C, CDKN2D, CCND1, CCND3, or CDKN1A was significantly associated with survival (P > 0.05). The trend tests were nominally significant for CCNE1-05, CCND2-10, CCND2-18, and CCND2-17 (P = 0.033, P = 0.043, P = 0.024, and P = 0.020, respectively; Table 2). The hazard associated with CCNE1-05 increased by 39% [per rare allele HR, 1.39; 95% confidence interval (95% CI), 1.04-1.85; Table 2]. CCND2-10 and CCND2-18 were correlated with CCND2-17 (r2 = 0.14 and 0.52, respectively). CCND2-10 was associated with reduced hazard (HR, 0.85; 95% CI, 0.73-1.00), whereas CCND2-18 and CCND2-17 were associated with increased hazard [HR, 1.14 (95% CI, 1.02-1.27) for CCND2-18; HR, 1.16 (95% CI 1.03-1.31) for CCND2-17; Table 2]. Figure 1 shows the cumulative hazard by genotype for the most significant association (CCND2-17). A model including all three significant CCND2 SNPs was significant (likelihood ratio test, P = 0.019). However, when using a forward stepwise regression procedure only, CCND2-18 was retained in the final model (P = 0.02). The haplotype analysis including these three CCND2 SNPs showed that the haplotype comprising the common allele of CCND2-10, the rare allele of CCND2-18, and the rare allele of CCND2-17 was associated with ovarian cancer survival [haplotype frequency, 0.25; P = 0.013; HR, 1.21 (95% CI, 1.04-1.41)] using the most common haplotype (comprising the common alleles of the three SNPs; frequency = 0.38) as baseline.
Table 2.
Genotype frequencies and results of univariate Cox regression analysis of common polymorphisms and ovarian cancer survival with P < 0.2
| Rs number | SNP ID | Genotype frequencies |
Trend test* | P | Risk per allele, HR (95% CI) | |||
|---|---|---|---|---|---|---|---|---|
| AA | Aa | aa | Total | χ2 | ||||
| rs3092904 | RB1-13 | 735 | 501 | 92 | 1,328 | 3.68 | 0.06 | 1.15 (1.00-1.31) |
| rs992519 | CDK6-07 | 1,095 | 363 | 29 | 1,487 | 2.11 | 0.15 | 0.89 (0.75-1.05) |
| rs445 | CDK6-06 | 1,207 | 270 | 14 | 1,491 | 2.06 | 0.15 | 1.15 (0.96-1.37) |
| rs3731249 | CDKN2A-12 | 1,401 | 70 | 3 | 1,474 | 1.71 | 0.19 | 0.80 (0.56-1.13) |
| rs11515 | CDKN2A-10 | 1,081 | 370 | 38 | 1,489 | 2.56 | 0.11 | 0.88 (0.75-1.03) |
| rs3217795 | CCND2-01 | 1,250 | 238 | 7 | 1,495 | 1.69 | 0.19 | 1.14 (0.94-1.38) |
| rs3217862 | CCND2-10 | 1,039 | 401 | 40 | 1,480 | 4.09 | 0.043 | 0.85 (0.73-1.00) |
| rs3217901 | CCND2-18 | 458 | 759 | 272 | 1,489 | 5.07 | 0.024 | 1.14 (1.02-1.27) |
| rs3217906 | CCND2-14 | 813 | 570 | 94 | 1,477 | 2.46 | 0.12 | 1.11 (0.98-1.26) |
| rs3217916 | CCND2-15 | 778 | 606 | 102 | 1,486 | 3.09 | 0.08 | 0.89 (0.78-1.02) |
| rs3217925 | CCND2-16 | 832 | 568 | 80 | 1,480 | 2.60 | 0.11 | 0.90 (0.78-1.03) |
| rs3217933 | CCND2-17 | 799 | 599 | 90 | 1,488 | 5.43 | 0.020 | 1.16 (1.03-1.31) |
| rs3217936 | CCND2-07 | 683 | 671 | 134 | 1,488 | 2.41 | 0.12 | 0.91 (0.80-1.03) |
| rs2069408 | CDK2-02 | 692 | 635 | 159 | 1,486 | 2.44 | 0.12 | 0.91 (0.81-1.03) |
| rs3218038 | CCNE1-05 | 1,374 | 112 | 3 | 1,489 | 4.55 | 0.033 | 1.39 (1.04-1.85) |
| rs762624 | CDKN1A-07 | 771 | 597 | 117 | 1,485 | 2.19 | 0.14 | 0.91 (0.81-1.03) |
| rs1410492 | CCND3-01 | 828 | 569 | 92 | 1,489 | 1.63 | 0.20 | 1.09 (0.96-1.24) |
| rs2069506 | CDK4-02 | 696 | 620 | 174 | 1,490 | 2.01 | 0.16 | 0.92 (0.82-1.03) |
| rs1420023 | CDKN1B-08 | 1,190 | 277 | 18 | 1,485 | 2.36 | 0.13 | 1.15 (0.97-1.36) |
Stratified by study.
NOTE: Data in bold highlight the statistically significant results.
Abbreviations: AA, common homozygote; Aa, heterozygote; aa, rare homozygote.
Fig. 1.
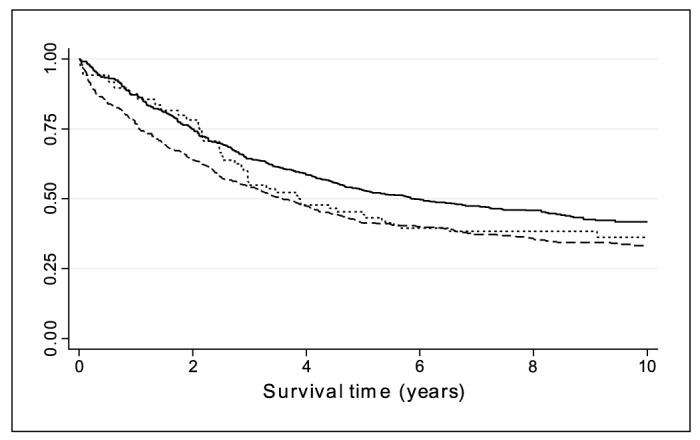
Cumulative survival (Kaplan-Meier plot) by CCND2 rs3217933 genotype. Solid line, common homozygote; dashed line, heterozygote; dotted line, rare homozygote.
Ten SNPs were tagged by a haplotype comprising the multi-SNPs. There was no association between survival and the multimarker haplotype (Table 3). Data on tumor stage, histopathology, grade, and age at diagnosis were available for 79%, 99.6%, 75%, and 100% of the cases, respectively (Table 1). These data were recorded and collected differently for each study, and so their completeness varies by study. In particular, the proportion of SEARCH patient with missing data on tumor stage and grade was higher, mostly because insufficient information was available in the medical record. As expected, each of these factors was significantly associated with outcome when considered independently (P < 0.05), but only age at diagnosis, stage, grade, and papillary NOS histologic subtype were significantly associated with survival in a multivariate analysis (data not shown). After adjusting for these factors, the HRs for CCND2-17 and CCND2-18 were not attenuated [Ptrend = 0.006; HR, 1.21 (95% CI, 1.06-1.39) for CCND2-17 and Ptrend = 0.003; HR, 1.21 (95% CI, 1.07-1.37) for CCND2-18]. CCNE1-05 was similar to not adjusted, but CCND2-10 was no longer significant for trend (Ptrend = 0.075). CDKN1B-02 was no longer significant on heterogeneity test (P = 0.47).
Table 3.
Cox regression analysis of multi-SNP haplotype–tagged SNPs and ovarian cancer survival
| Gene | Tagging SNPs | Haplotype* | Tagged SNPs | Missing data† | Haplotype frequency | HR (95% CI) | P |
|---|---|---|---|---|---|---|---|
| CDK6 | rs4729049, rs992519 | 11 | rs2374594, rs6975474, rs2374589, rs10246604 | 2 | 0.068 | 1.00 (0.75-1.32) | 1.0 |
| rs3217862, rs3217820 | 00 | rs3217827 rs3217881 | 3 | 0.48 | 1.00 (0.88-1.14) | 1.0 | |
| rs3217869, rs3217852 | 00 | rs3217830 rs3217896 | 2 | 0.37 | 1.00 (0.87-1.15) | 1.0 | |
| CCND2 | rs3217926, rs3217925, rs3217916 | 000 | rs3217907 | 7 | 0.35 | 1.00 (0.87-1.15) | 1.0 |
| rs3217926, rs3217936 | 00 | rs4625554 | 5 | 0.30 | 1.00 (0.86-1.19) | 1.0 |
0 represents common allele of the tagging SNP in the haplotype; 1 represents rare allele of the tagging SNP in the haplotype.
Number of subjects missing 50% or more of the SNP data for each haplotype.
The P values presented above have not been adjusted for multiple hypothesis testing. Because SNPs within the same gene may be in linkage disequilibrium, the test statistics are not independent, and standard methods for adjusting for multiple testing, such as the Bonferroni correction, would be too conservative. We therefore used simulations to estimate empirical P values adjusted for multiple testing by randomly shuffling the survival time and outcome among individuals' multiple times, and estimated how frequently a P value as extreme as those observed were obtained from the randomly permuted data. After adjusting for multiple testing, the P value for the most significant association (CCND2-17, P = 0.020) was 0.68, suggesting that these findings are likely to be the result of chance.
Discussion
The biological factors involved in the development and progression of ovarian cancer are poorly understood. We have evaluated the effect of 97 SNPs in 14 cell cycle genes on ovarian cancer survival among White women from the East Anglia and West Midland regions of the United Kingdom, United States, and Denmark. We have previously found polymorphisms in CDKN2A/CDKN2B, RB1, and CCND1 to be marginally associated with ovarian cancer risk (26, 27). However, no published studies have systematically investigated common variation in these genes and survival after a diagnosis of ovarian cancer. The major strengths of this study are its large sample size, the fact that its component studies are population based, the length and systematic approach of the follow-up, and the systematic approach to tagging the known common genetic variation in the genes of interest.
The differences between the component studies are a potential weakness, but these differences are unlikely to bias the results or influence their interpretation. The time between diagnosis and recruitment was longer for SEARCH than for the other studies, and so the cancers tended to have better prognostic features (nonserous subtype, lower grade, and earlier stage) and the average annual mortality rate was lower. However, we allowed for this in the analysis, and the estimates of the genotype effects will not be biased, provided that the Cox proportional hazards assumption is not violated. There are also likely to be differences in the way the patients were treated by study center, but again this will not result in any bias as treatment is unlikely to be correlated with individual genotype.
We have found no evidence that common variations in RB1, CDK2, CDK4, CDK6, CDKN2A, CDKN2B, CDKN2C, CDKN2D, CCND1, CCND3, or CDKN1A are associated with outcome after a diagnosis of ovarian cancer. We observed marginally significant evidence for an association between survival and SNPs in CCND2, CDKN1B, and CCNE1, but these results were not significant after adjusting for multiple hypothesis testing. The SNPs under study were selected to adequately tag all the common variants in each gene, and not because of their predicted effects on structure and function. Nevertheless, it is possible that important, unidentified variants were not efficiently tagged. Furthermore, some known common variants were poorly tagged because of tag SNP assay failure. Again, power to detect association with these SNPs is limited. It is also possible that rare variants in these genes are important predictors of outcome. For these genes, it is likely that EGP resequencing and Hapmap data sets identified most of the common variants, and we are confident that the set of tag SNPs we selected has adequately tagged the known and unknown common variants within each gene. We may also have failed to detect any association with survival because of lack of statistical power to detect modest effects. Despite our large sample size, there were just 637 deaths in our cohort. Assuming a type I error rate of 0.05, we had only 51% power to detect a codominant allele of frequency of 0.1 that confers a relative hazard of 1.3 and 86% power to detect a similar allele with frequency of 0.3. Power to detect recessive alleles with similar effects was poor. Power may also be reduced by the use of all-cause mortality rather than ovarian cancer–specific mortality as an end point because some women will have died from other causes that might not be related to genotype. However, in the age group of women included in these studies, the proportion of women dying from causes unrelated to ovarian cancer is likely to be small and any reduction in power will be limited.
In conclusion, common variation in the 14 genes of cell cycle pathways does not seem to be associated with moderate variation in prognosis after a diagnosis of ovarian cancer. Much larger studies will be needed to exclude common variants with small effects.
Supplementary Material
Acknowledgments
We thank Hannah Munday, Barbara Perkins, Clare Jordan, Judy West, Anabel Simpson, Sue Irvine, the local general practices and nurses, and the East Anglian Cancer Registry for recruitment of the U.K. cases; the EPIC-Norfolk investigators for recruitment of the U.K. controls; Danielle Shadforth, Jonathan Tyrer, Craig Luccarini, and Don Conroy for expert technical assistance; and finally, all the study participants who contributed to this research.
Grant support: Cancer Research UK, the Roswell Park Alliance, the Danish Cancer Society, and the National Cancer Institute grant CA71766 and Core Grants CA16056 and RO1 CA61107; a grant from WellBeing of Women (H. Song); and the Mermaid component of the Eve Appeal (S.J. Ramus). S.A. Gayther is a Higher Education Funding Council for England – funded senior lecturer. D.F. Easton is a Principal Research Fellow and P.D.P. Pharoah is a Senior Clinical Research Fellow of Cancer Research UK.
Footnotes
Publisher's Disclaimer: The costs of publication of this articlewere defrayed in part by the payment of page charges.This article must therefore be hereby marked advertisement in accordance with18 U.S.C. Section 1734 solely to indicate this fact.
Note: Supplementary data for this are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Ben DY, Chetrit A, Hirsh-Yechezkel G, et al. Effect of BRCA mutations on the length of survival in epithelial ovarian tumors. J Clin Oncol. 2002;20:463–6. doi: 10.1200/JCO.2002.20.2.463. [DOI] [PubMed] [Google Scholar]
- 3.Majdak EJ, Debniak J, Milczek T, et al. Prognostic impact of BRCA1 pathogenic and BRCA1/BRCA2 unclassified variant mutations in patients with ovarian carcinoma. Cancer. 2005;104:1004–12. doi: 10.1002/cncr.21276. [DOI] [PubMed] [Google Scholar]
- 4.Gadducci A, Di Cristofano C, Zavaglia M, et al. P53 gene status in patients with advanced serous epithelial ovarian cancer in relation to response to paclitaxel plus platinum-based chemotherapy and long-term clinical outcome. Anticancer Res. 2006;26:687–93. [PubMed] [Google Scholar]
- 5.Hefler LA, Mustea A, Konsgen D, et al. Vascular endothelial growth factor gene polymorphisms are associated with prognosis in ovarian cancer. Clin Cancer Res. 2007;13:898–901. doi: 10.1158/1078-0432.CCR-06-1008. [DOI] [PubMed] [Google Scholar]
- 6.Spurdle AB, Hopper JL, Chen X, et al. The steroid 5α-reductase type II TA repeat polymorphism is not associated with risk of breast or ovarian cancer in Australian women. Cancer Epidemiol Biomarkers Prev. 2001;10:1287–93. [PubMed] [Google Scholar]
- 7.Beeghly A, Katsaros D, Chen H, et al. Glutathione S-transferase polymorphisms and ovarian cancer treatment and survival. Gynecol Oncol. 2006;100:330–7. doi: 10.1016/j.ygyno.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 8.Green H, Soderkvist P, Rosenberg P, Horvath G, Peterson C. mdr-1 single nucleotide polymorphisms in ovarian cancer tissue: G2677T/A correlates with response to paclitaxel chemotherapy. Clin Cancer Res. 2006;12:854–9. doi: 10.1158/1078-0432.CCR-05-0950. [DOI] [PubMed] [Google Scholar]
- 9.Hefler LA, Grimm C, Ackermann S, et al. An interleukin-6 gene promoter polymorphism influences the biological phenotype of ovarian cancer. Cancer Res. 2003;63:3066–8. [PubMed] [Google Scholar]
- 10.Higashi T, Kyo S, Inoue M, Tanii H, Saijoh K. Novel functional single nucleotide polymorphisms in the latent transforming growth factor-β binding protein-1L promoter: effect on latent transforming growth factor-β binding protein-1L expression level and possible prognostic significance in ovarian cancer. J Mol Diagn. 2006;8:342–50. doi: 10.2353/jmoldx.2006.050133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hogdall EV, Kjaer SK, Glud E, et al. Evaluation of a polymorphism in intron 2 of the p53 gene in ovarian cancer patients. From the Danish “Malova” Ovarian Cancer Study. Anticancer Res. 2003;23:3397–404. [PubMed] [Google Scholar]
- 12.Obata H, Yahata T, Quan J, Sekine M, Tanaka K. Association between single nucleotide polymorphisms of drug resistance-associated genes and response to chemotherapy in advanced ovarian cancer. Anticancer Res. 2006;26:2227–32. [PubMed] [Google Scholar]
- 13.Santos AM, Sousa H, Portela C, et al. TP53 and P21 polymorphisms: response to cisplatinum/paclitaxel-based chemotherapy in ovarian cancer. Biochem Biophys Res Commun. 2006;340:256–62. doi: 10.1016/j.bbrc.2005.11.176. [DOI] [PubMed] [Google Scholar]
- 14.Six L, Grimm C, Leodolter S, et al. A polymorphism in the matrix metalloproteinase-1 gene promoter is associated with the prognosis of patients with ovarian cancer. Gynecol Oncol. 2006;100:506–10. doi: 10.1016/j.ygyno.2005.08.049. [DOI] [PubMed] [Google Scholar]
- 15.Kang S, Ju W, Kim JW, et al. Association between excision repair cross-complementation group 1 polymorphism and clinical outcome of platinum-based chemotherapy in patients with epithelial ovarian cancer. Exp Mol Med. 2006;38:320–4. doi: 10.1038/emm.2006.38. [DOI] [PubMed] [Google Scholar]
- 16.Li AJ, Lerner DL, Gapuzan ME, Karlan BY. AIB1 polymorphisms predict aggressive ovarian cancer phenotype. Cancer Epidemiol Biomarkers Prev. 2005;14:2919–22. doi: 10.1158/1055-9965.EPI-05-0540. [DOI] [PubMed] [Google Scholar]
- 17.Pinto D, Pereira D, Portela C, da Silva JL, Lopes C, Medeiros R. The influence of HER2 genotypes as molecular markers in ovarian cancer outcome. Biochem Biophys Res Commun. 2005;335:1173–8. doi: 10.1016/j.bbrc.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 18.Pharoah PD, Dunning AM, Ponder BA, Easton DF. Association studies for finding cancer-susceptibility genetic variants. Nat Rev Cancer. 2004;4:850–60. doi: 10.1038/nrc1476. [DOI] [PubMed] [Google Scholar]
- 19.Elledge SJ. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–72. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 20.Hashiguchi Y, Tsuda H, Inoue T, Nishimura S, Suzuki T, Kawamura N. Alteration of cell cycle regulators correlates with survival in epithelial ovarian cancer patients. Hum Pathol. 2004;35:165–75. doi: 10.1016/j.humpath.2003.07.018. [DOI] [PubMed] [Google Scholar]
- 21.Betticher DC, Thatcher N, Altermatt HJ, Hoban P, Ryder WD, Heighway J. Alternate splicing produces a novel cyclin D1 transcript. Oncogene. 1995;11:1005–11. [PubMed] [Google Scholar]
- 22.Matthias C, Branigan K, Jahnke V, et al. Polymorphism within the cyclin D1 gene is associated with prognosis in patients with squamous cell carcinoma of the head and neck. Clin Cancer Res. 1998;4:2411–8. [PubMed] [Google Scholar]
- 23.Dhar KK, Branigan K, Howells RE, et al. Prognostic significance of cyclin D1 gene (CCND1) polymorphism in epithelial ovarian cancer. Int J Gynecol Cancer. 1999;9:342–7. doi: 10.1046/j.1525-1438.1999.99048.x. [DOI] [PubMed] [Google Scholar]
- 24.Shu XO, Moore DB, Cai Q, et al. Association of cyclin D1 genotype with breast cancer risk and survival. Cancer Epidemiol Biomarkers Prev. 2005;14:91–7. [PubMed] [Google Scholar]
- 25.Kommoss S, du BA, Ridder R, et al. Independent prognostic significance of cell cycle regulator proteins p16(INK4a) and pRb in advanced-stage ovarian carcinoma including optimally debulked patients: a translational research subprotocol of a randomised study of the Arbeitsgemeinschaft Gynaekologische Onkologie Ovarian Cancer Study Group. Br J Cancer. 2007;96:306–13. doi: 10.1038/sj.bjc.6603531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song H, Ramus SJ, Shadforth D, et al. Common Variants in RB1 Gene and Risk of Invasive Ovarian Cancer. Cancer Res. 2006;66:10220–6. doi: 10.1158/0008-5472.CAN-06-2222. [DOI] [PubMed] [Google Scholar]
- 27.Gayther SA, Song H, Ramus SJ, et al. Tagging single nucleotide polymorphisms in cell cycle control genes and susceptibility to invasive epithelial ovarian cancer. Cancer Res. 2007;67:3027–35. doi: 10.1158/0008-5472.CAN-06-3261. [DOI] [PubMed] [Google Scholar]
- 28.Lum A, Le Marchand L. A simple mouthwash method for obtaining genomic DNA in molecular epidemiological studies. Cancer Epidemiol Biomarkers Prev. 1998;7:719–24. [PubMed] [Google Scholar]
- 29.Zaykin DV, Westfall PH, Young SS, Karnoub MA, Wagner MJ, Ehm MG. Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Hum Hered. 2002;53:79–91. doi: 10.1159/000057986. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


