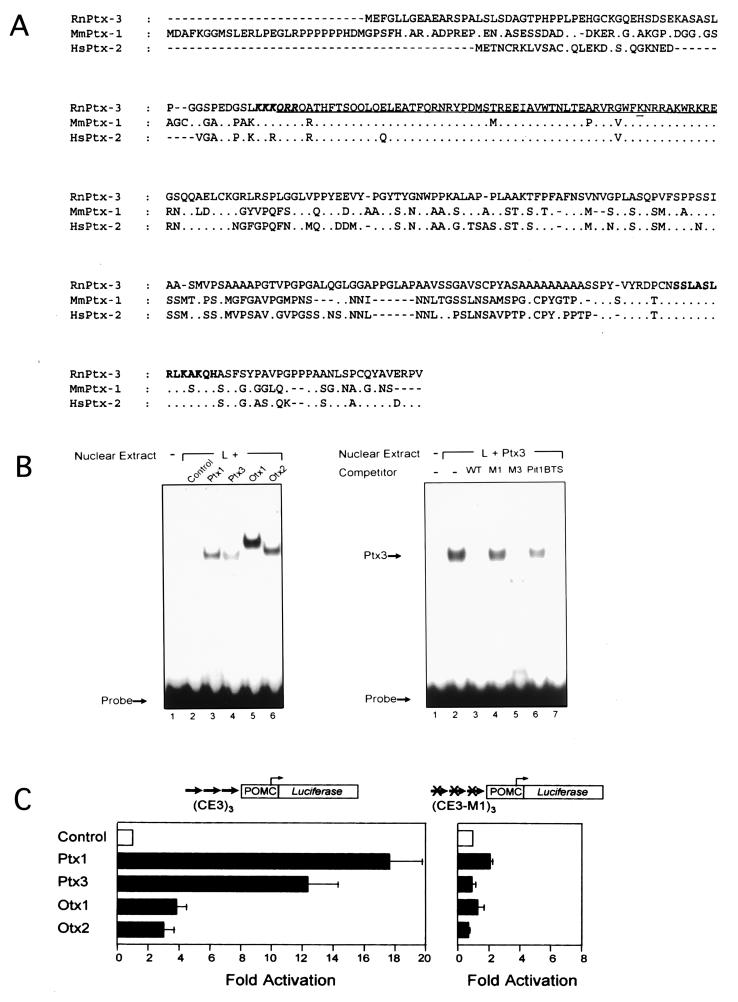
Figure 1.
Structure and properties of PTX3. (A) Primary structure of Ptx3 deduced from the ORF of the Ptx3 cDNA and alignment to Ptx1 (11–13) and Ptx2 (RGS) (14). The homeodomain is underlined, with the bicoid-type-specific lysine at position 9 of the third helix double-underlined (23). A consensus nuclear localization signal is shown in boldface, italic type (24). The conserved C- terminal domain (14) is in boldface type. (B) Binding of Ptx3 to the CE3 element of the rat POMC promoter in gel shift analysis. Ptx3 binds to the CE3 element of the rat POMC promoter as was found for the related factors Ptx1, Otx1, and Otx2. Competition experiments using mutant recognition sites (11) within (M1) and outside (M3) the bicoid core of the CE3 element (200-fold molar excess) show that the ineffective mutant M3 and the wild type compete for Ptx3 binding, because the effective mutant M1 cannot. (C) Transactivational properties of ptx3 in transient transfection assays. The activity of Ptx3 on an artificial promoter construct based on three copies of the POMC-CE3 element is comparable to that of Ptx1 (11), whereas Otx1 and Otx2 are clearly less active. When using the M1 mutated binding site, the activity is lost.