Figure 3.
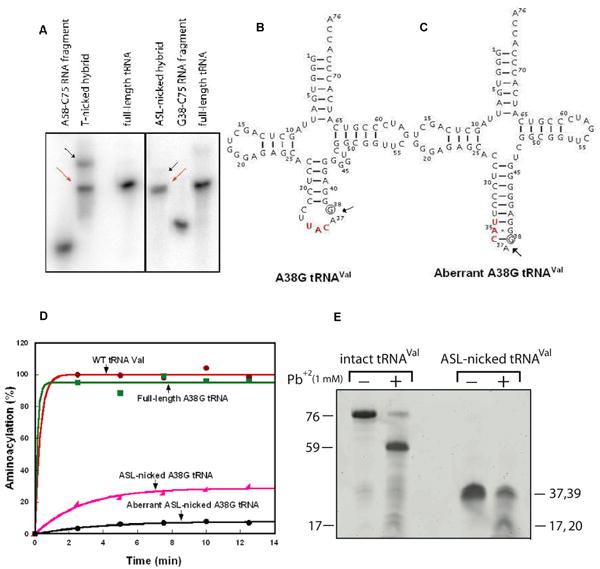
Characterization of nicked tRNAs. (A) Formation of the T-nicked (left) and ASL-nicked (right) tRNAC75 substrates by hybridization of an unlabeled 5′-fragment with a 32P-labeled 3′ fragment. Full-length and intact 32P-tRNAC75 was run as a marker. The nicked substrates (indicated by the red arrows) that co-migrated with the intact tRNA marker were isolated from gels and used for analysis of CCA addition, while the aberrant hybridization products that migrated slower than the intact tRNA were marked by black arrows and were not further analyzed for CCA addition. (B) Sequence and cloverleaf structure of the A38G mutant of E. coli tRNAVal, where the A38G substitution is indicated by a circle. (C) The aberrant structure of the ASL-nicked A38G tRNAVal. (D) Aminoacylation of tRNAVal transcripts (2 μM) by purified E. coli ValRS (4 μM). (E) Lead cleavage of intact A38G tRNA and the ASL-nicked A38G tRNA analyzed by a 12% denaturing PAGE. The numbers on the sides of the gel indicate the lengths of tRNA or fragments.
