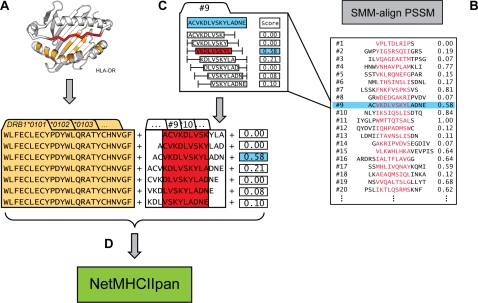
Figure 1. Schematic Illustration of the NetMHCIIpan Method.
(A) The HLA-DR pseudo sequence is constructed from polymorphic HLA-DR residues in potential contact with a bound peptide. (B) Position specific scoring matrix (PSSM) and peptide core alignment (shown in red) is made for each allele using the SMM-align method [26]. N and C terminal peptide flanking regions, PFR, are identified as the up to three amino acids flanking the peptide-binding core. (C) Suboptimal peptides are presented to the NetMHCpan method with binding values normalized to the optimal peptide score (for the peptide shown in red) as described in Materials and Methods. (D) The NetMHCIIpan method is trained integrating data from all alleles. Input to the artificial neural network training includes the peptide core, composition and length of the N and C terminal PFR, length of the source peptide as well as the normalized binding affinity value (for details see Materials and Methods).