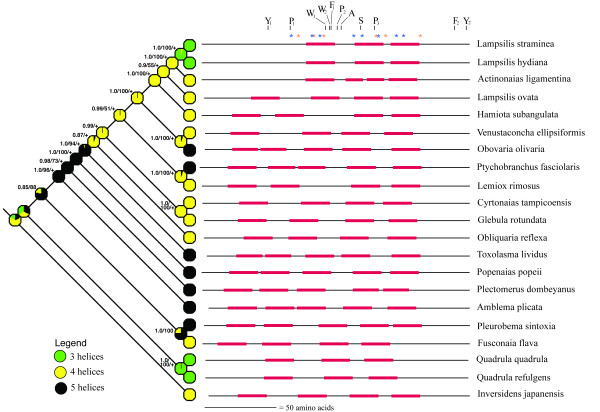
Figure 2.
Schematic representation of 21 unionoidean bivalve MCOX2 C-terminus extensions and their evolutionary relationships. A Bayesian Inference estimate of phylogeny (with posterior probabilities [via Mr. Bayes], maximum parsimony bootstrap percentages [when >50%; via PAUP*], and a "+" to indicate significant maximum likelihood ancestral state reconstructions [via Mesquite] given at the tree nodes), based on analyses of 2310 unambiguously alignable protein coding nucleotides from the F and Mcox2-cox1 gene junction regions, and a maximum likelihood optimization of TMH number are presented. The black horizontal lines are proportional to extension length and the red rectangles represent predicted TMH locations and lengths (via ConPredII). The 11 constant amino acid positions are indicated by single letter codes; the eight HyPhy and six codeml sites potentially inferred to be under positive selection are indicated by blue and orange asterisks, respectively (also see Table 2; see Additional file 1).