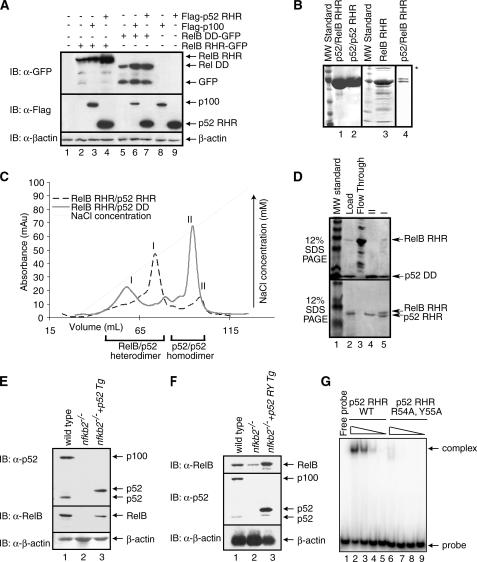
FIGURE 2.
RelB RHR stabilization by p52 in vitro. A, Western blot analysis of the steady state levels of RelB RHR and RelB DD proteins in HEK 293 cells transfected with FLAG-p100, FLAG-p52, and RelB-GFP in different combinations. In the presence of p100 and p52, RelB protein levels are enhanced. B, Coomassie-stained SDS-PAGE showing purity of p52-RelB heterodimer, p52 homodimer, and RelB RHR homodimer. A lower concentration of p52-RelB heterodimer is shown in lane 4 to visualize the separation between the two proteins. RelB continuously degrades during purification, and the degradation products are seen in the gel (lane 4). Aggregated species of RelB in lane 3 is marked by an asterisk. C, co-refolded mixtures of RelB RHR and p52 RHR (black), and RelB RHR and p52 DD (gray) were separated by cation exchange (S-Sepharose column) chromatography. D, Coomassie-stained SDS-PAGE of samples from the S column chromatography of the RelB RHR-p52 DD (top panel) and RelB RHR-p52 RHR (bottom panel). The load appears to contain excess p52, which masks the RelB RHR. E, Western blot analysis of the steady state levels of p100/p52 (top panel) and RelB (middle panel) in wt, nfkb2-/- MEF and p52-reconstituted nfkb2-/- cells. Reconstituted p52 migrates higher due to the exact site of processing of p100 is unknown. F, DNA binding-defective p52 mutant stabilizes RelB. Western blot analysis of the steady state levels of p100/p52 (top panel) and RelB (middle panel) in wt, nfkb2-/- MEF, and p52-reconstituted nfkb2-/- cells. Reconstituted p52 migrates higher due to the exact site of processing of p100 is unknown. G, electrophoretic mobility shift assay analysis of wild type p52 and p52 R54A,Y55A double mutant. Lanes 2 and 6, 3 and 7, 4 and 8, and 5 and 9 contain 1100, 250, 25, and 2.5 nm wild type and double mutant p52, respectively.