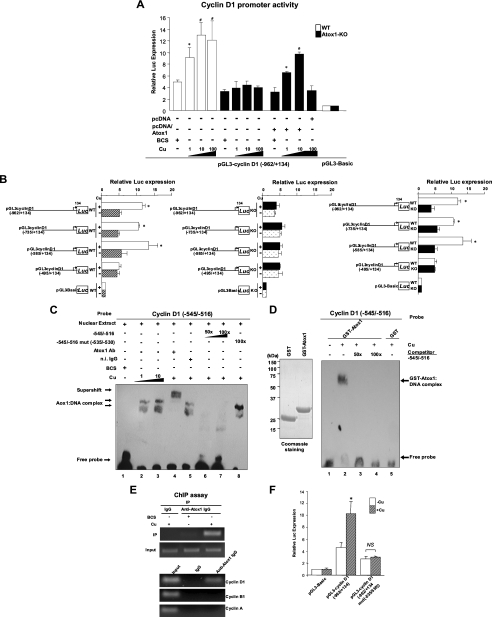
FIGURE 3.
Atox1 binds to and activates the cyclin D1 promoter in a copper-dependent manner. A, effect of copper on transactivation of the cyclin D1 gene promoter in WT and Atox1-KO MEFs. Cells were transiently transfected with cyclin D1 promoter luciferase reporter constructs (pGL3-cyclin D1 (-962/+134)) or empty reporter constructs (pGL3-Basic) along with either pcDNA/Atox1 or pcDNA. Cells were treated with either BCS (200 μm) or CuCl2 at the dose indicated. Two days after transfection, the luciferase activity was assayed and normalized to the Renilla luciferase activity produced by the co-transfected control plasmid pRL-CMV. Results shown are means ± S.E. from at least three independent transfection experiments, each performed in quadruplicate (*, p < 0.01; #, p < 0.001 versus BCS-treated WT cells, or BCS-treated Atox1-/- cells transfected with pcDNA/Atox1). B, identification of copper/Atox1-responsive elements in a proximal 90-bp cyclin D1 promoter element. WT and Atox1-KO MEFs were transiently transfected with 5′ deletion constructs of cyclin D1 in the presence of either CuCl2 (10 μm, +Cu) or BCS (200 μm, -Cu). Left and middle panels, relative luciferase activity in WT or Atox1-KO MEFs in the presence of either CuCl2 (10 μm, +Cu) or the copper chelator BCS (200 μm, -Cu). Right panel, relative luciferase activity in wild-type and Atox1-/- cells in the presence of CuCl2 (10 μm). Results shown are means ± S.E. from at least three independent transfection experiments, each performed in quadruplicate (*, p < 0.01 versus BCS-treated WT (left panel), or CuCl2-treated Atox1-/-cells (right panel)). C and D, EMSA, showing the binding of Atox1 to the region -535 to -530 in the cyclin D1 promoter in a copper-dependent manner. C, nuclear extracts from MEFs were incubated with the biotinylated cyclin D1 promoter fragment with indicated treatments. D, left panel shows purified GST and GST-Atox1. Right panel, purified GST or GST-Atox1 was incubated with the DNA probe with indicated treatments. E, ChIP assay showing association of Atox1 with the cyclin D1 promoter in a copper-dependent manner in vivo. Cells were treated with either indicated treatments (upper panel) or CuCl2 (10 μm) (lower panel) and cross-linked with 1% formaldehyde. Nuclear lysates were immunoprecipitated (IP) with anti-Atox1 antibody or normal IgG, and the promoter region of either cyclin D1, cyclin B1, or cyclin A was amplified by PCR. A small aliquot of lysates before IP were used for PCR amplification as the input control (Input). Results are representative of three independent experiments. F, region -535 to -530 is required for copper-induced activation of the cyclin D1 promoter. MEFs were transfected with a cyclin D1 promoter luciferase reporter construct (pGL3-cyclin D1 (-962/+134)) with or without mutation of the copper/Atox1-responsive element (-535 to -530 region). *, p < 0.01 versus BCS-treated cells.