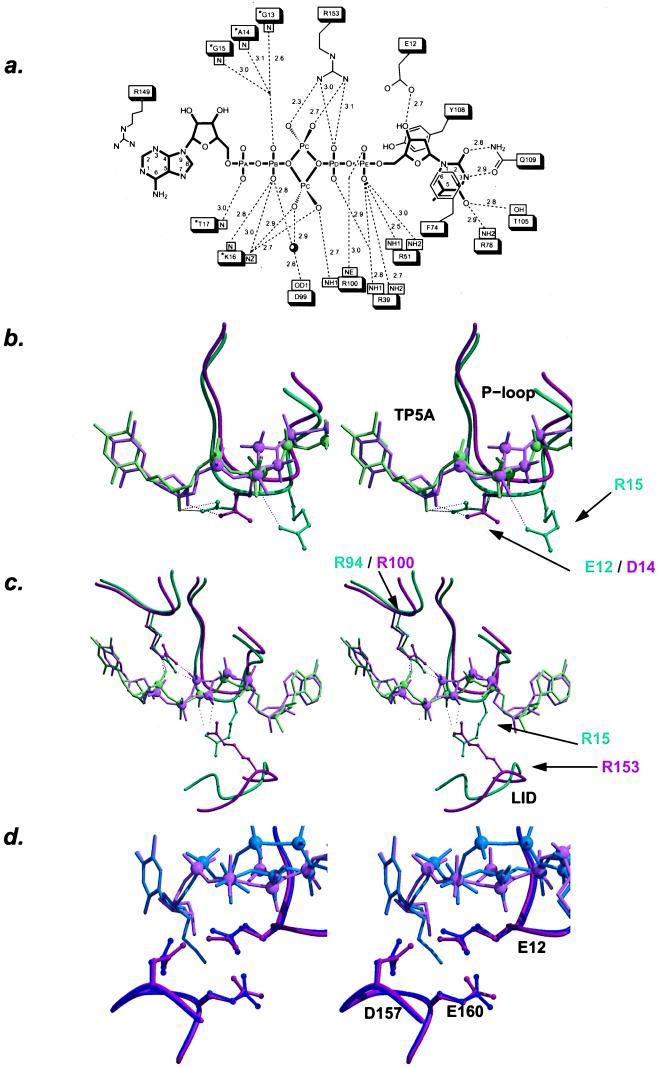
Figure 1.
Interactions of the bisubstrate inhibitor with TmpK (a). Distance map of TP5A bound to TmpKcoli. P loop residues are marked with an asterisk. (b–d) Stereoviews. Overlay of the TmpKcoli–TP5A complex model (pink) with the TmpKyeast–TP5A model (green) (b and c) or the TmpKcoli–AZTP5A (blue) (d). (b) Interactions of the 3′-hydroxyl of the thymidine deoxyribose. In TmpKyeast, a bidentate interaction between the P loop aspartic acid and the sugar hydroxyl is observed. The binding of AZT-MP causes the P loop to move, thus displacing the catalytic P loop arginine. In contrast, in TmpKcoli, the interaction between Glu-12 and the 3′-hydroxyl is side-on, and the bulkier azido group does not induce a significant movement of the P loop. (c) Similar phosphate–arginine interactions made in TmpKyeast by Arg-15 and in TmpKcoli by Arg-153. Displayed are the P loop and a part of the Lid region. The structures were overlaid according to the position of the bisubstrate inhibitor. (d) In the TmpKcoli–TP5A and the TmpKcoli–AZTP5A complex structures, the thymine base is at an identical position, but the deoxyribose moiety has undergone a rigid-body rotation caused by the azido group in the AZT-P5A complex. In addition, Glu-12 has rotated slightly to provide more room for the azido group. The rotation of the deoxyribose induces a similar rotation of the Glu-160 side chain. As Glu-12 makes close interactions with Asp-157, the latter carboxylic acid also rotates slightly. b–d were generated by using bobscript (29, 30) and raster 3d (31).