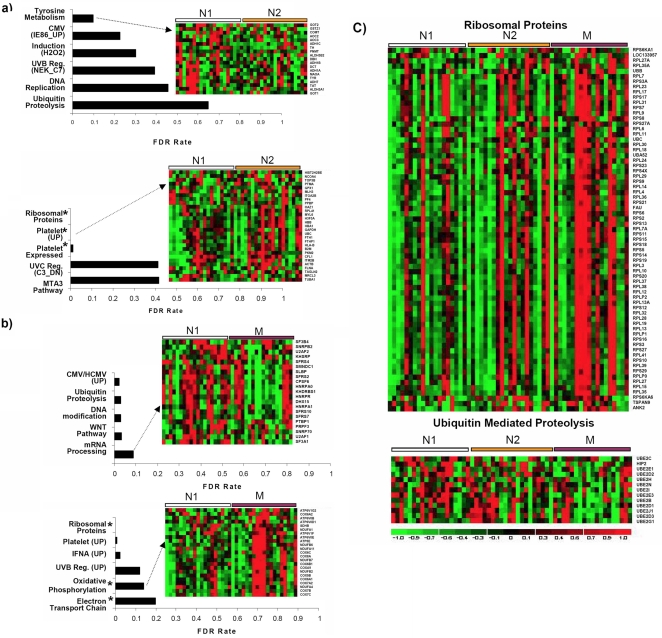
Figure 2. GSEA Analysis.
The analysis has been performed for >1200 predefined datasets using GSEA 2.0 software. Signal values for each gene are obtained by collapsing the probe values using max_probe algorithm. Representative datasets, significantly enriched (FDR<50%, or NPV< = 0.01) between any two groups and corresponding heatmaps (depicting relative gene expression changes of core enrichment) are shown in a) N2 vs. N1 and b) M vs. N1. Datasets that are enriched in both the original and validation analyses are marked with *. c) Heatmaps of ribosomal proteins and ubiquitin mediated proteolysis illustrate transitional trends in gene expression across the N1, N2 and M groups.