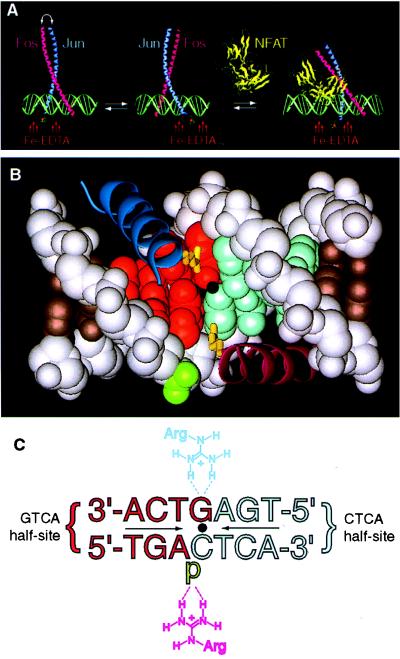
Figure 1.
The stereochemistry of DNA-bound Fos and Jun alone and in the presence of NFAT. (A) AP-1 alone binds DNA as a mixture of two stereoisomers, which are related by subunit interchange (left and center). In the cooperative complex formed by AP-1 and NFAT on DNA (right), Fos•Jun is locked into a single stereochemical orientation. The stereochemistry of the protein•DNA complex is reported by a covalently attached Fe-EDTA moiety, which effects proximity-directed hydroxyl radical cleavage of DNA. The model is derived from the x-ray structure of AP-1•DNA (9) and NFATp•AP-1•DNA (34). (B) Contacts between the basic regions of Fos•Jun central base pair of a consensus AP-1 site (9). In the orientation shown, Jun (dark blue α-helix) is bound over the GTCA half-site (red space-filling model), and its residue Arg-279 (yellow tube) makes a base-specific bidentate hydrogen bond to the central guanine; Fos (purple α-helix) is bound over the CTCA half-site (light blue space-filling model), and its residue Arg-155 makes a nonspecific contact to the phosphate flanking the central cytosine (green). In the other orientational isomer (not shown), the locations of Fos and Jun are swapped, and the roles of Arg-155 and Arg-279 are interchanged. Dot denotes the center of pseudodyad symmetry. (C) Schematic representation of the contacts shown in Fig. 1B (p, phosphate). Coloring and orientation matched those of Fig. 1B. Note: the strand sense in Fig. 1C is written opposite to the usual convention (i.e., the sequence of the top strand is written in the 3′–5′ orientation going from left to right) to facilitate comparison of Figs. 1B and C; all other figures present the sequence in the conventional sense.