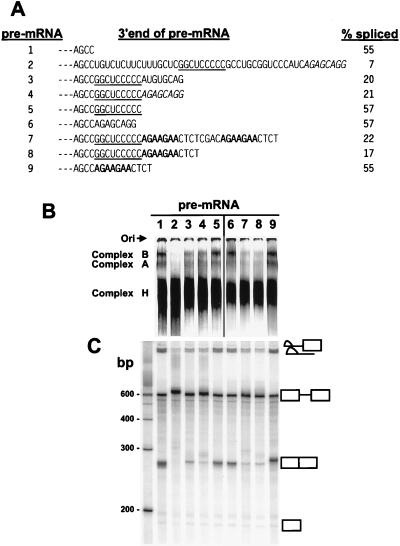
Figure 4.
Inhibition of BPV-1 late pre-mRNA splicing by the functional C-rich core of the ESS. (A) 3′ end sequences of the different pre-mRNAs that were used for in vitro spliceosome assembly (B) and splicing (C). The splicing efficiency of each pre-mRNA was calculated based on the gel profile in C. Small dashes on the left represent the sequences (including SE1) upstream. The functional C-rich core of the ESS is underlined. The ASF/SF2 binding site from the 3′ AG-rich part of the ESS and an optimal ASF/SF2 binding site octamer A3 (9) are indicated by italics and bold, respectively. Pre-mRNAs are identified at the left by the numbers 1–9. (B) Formation of spliceosome complexes on different pre-mRNA substrates with varied 3′ ends as shown in A. Spliceosome complex formation was performed under standard splicing condition (13, 22), stopped at 45-min incubation at 30°C by addition of heparin, and loaded and run immediately on a native 4% polyacrylamide gel. Spliceosome complexes A and B and the nonspecific complex H are indicated on the left of the gel. Lanes 1–5 were overexposed to compensate for loading variations of each pre-mRNA and to facilitate comparison of pre-mRNAs 3–5. Ori, origin. (C) Splicing gel electrophoresis of each correspondent pre-mRNA in B. The identity of the spliced products and intermediates is shown at the right. Positions of DNA size markers corresponding to a 100-bp ladder are shown at left of the gel.