Figure 1.
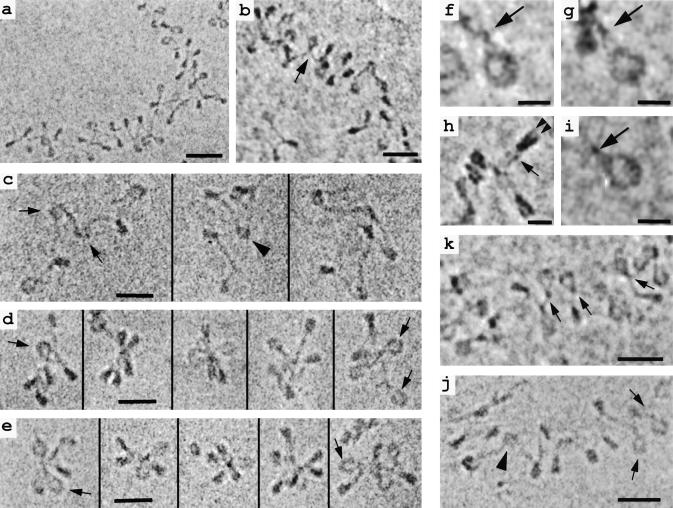
(a, b, and k) Soluble chromatin from chicken erythrocyte nuclei vitrified in ≈5 mM M+ and imaged unfixed and unstained in the frozen hydrated state. Nucleosomes and linker DNA are seen in many different orientations in these projections of the 3D structure. Arrows in b and k denote nucleosomes with the linker histone-dependent “stem” conformation described in the text. (c, d, and f–i) EC-M of unstained, unfixed chromatin reconstituted onto tandem DNA sequences containing the nucleosome-positioning sequence of 5S rDNA (30) and vitrified in 10 mM M+. All samples except c contain linker histone H5. En face views of nucleosomes (f, g, and i) show the linker DNA entering and exiting the nucleosome tangentially, then “intersecting” and remaining apposed for 3–5 nm before diverging (arrows). Edge-on views (h) show the two gyres of DNA (arrowheads) and the apposition of linker DNA (arrow). (c) Reconstituted hexanucleosomes without histone H5. In most nucleosomes, the linker DNA segments diverge after leaving the histone core (arrows), but in some cases they appear to “cross over” (arrowhead). (d) Reconstituted hexanucleosomes as in c, but with added linker histone H5. The chromatin particles are more compact and adopt a star-like conformation in which “stem” structures are common (arrows). (e) EC-M images of small oligonucleosomes released from chicken erythrocyte nuclei after micrococcal nuclease digestion. A zigzag, star-like conformation is seen, and, like the reconstituted hexanucleosomes in d, linker DNA segments show the “stem” architecture (arrows). (j) Chromatin released from COS-7 cells and vitrified in 20 mM M+ again shows a 3D zigzag conformation. While most nucleosomes show the stem motif (arrows), a few (arrowhead) have a linker DNA conformation typical of H1-free chromatin. [Bars = 10 nm (f–i); 30 nm for others.]