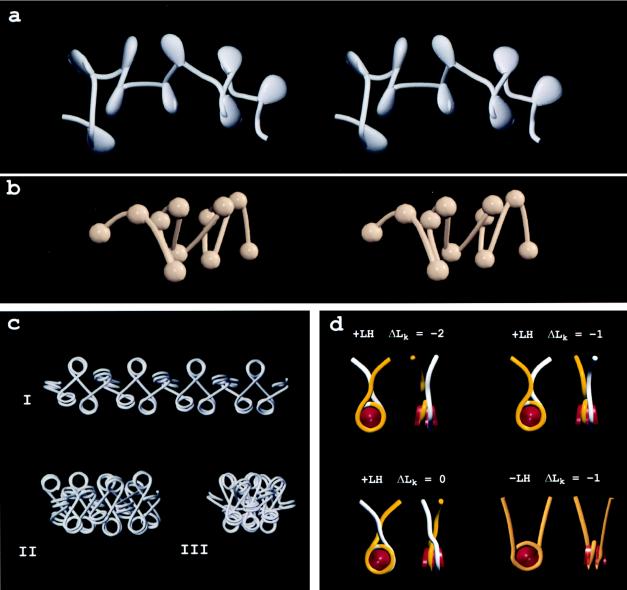
Figure 3.
(a) Stereo pair of a 3D model of a 9-nucleosome segment of a chicken erythrocyte chromatin fiber imaged in ≈5 mM M+. Nucleosomes and their associated “stems” are represented by pear-shaped solids, all of which point toward the fiber interior. (b) Stereo pair of a chicken erythrocyte oligonucleosome vitrified in 80 mM M+ and reconstructed from a tomographic tilt series. The quality of the reconstruction allowed nucleosome locations and linker paths to be identified, but details of nucleosome orientation and linker entry–exit sites were not resolved [Horowitz et al. (34)]. A movie of the reconstructed volume on which the model is based is available at http://www.ummed.edu/pub/r/rhorowit. (c) Models of uniform chromatin fibers based on principles discussed in Woodcock et al. (52), using the mean linker entry–exit angles measured from micrographs and reconstructions. Angles were 85° (I), 45° (II), and 34° (III). (d) Space-filling models of nucleosomes in the presence (+LH) and absence (−LH) of linker histones shown en face and in side views. With the stem conformation, differing linker paths, which appear to be energetically similar, result in differing linking numbers, ΔLk.