Abstract
Purpose
Pediatric cataract is the most common form of treatable childhood blindness and is both clinically and genetically heterogeneous. Autosomal dominant and recessive forms of cataract have been reported to be caused by mutations in 22 different genes so far. Of the cataract mutations reported to date, about half the mutations occur in crystallins, a quarter of the mutations in connexins, and the remainder is evenly divided between intrinsic membrane proteins, intermediate filament proteins, and transcription factors. This study is aimed at identification of the spectrum and frequency of crystallin gene mutations in cataractous patients in an Indian population.
Methods
Genetic analysis was extended to screen the entire coding region of the CRYAA, CRYAB, CRYBA1, CRYBA4, CRYBB1, CRYBB2, CRYBB3, CRYGC, CRYGD, and CRYGS genes using single stranded conformational polymorphism (SSCP) analysis as a screening technique followed by direct sequencing of all subjects that displayed an electrophoretic shift.
Results
This report describes the first simultaneous mutation analysis of 10 crystallin genes in the same population, represented by 60 south Indian families. The analysis allowed the identification of causative mutations in 10 of the families (three novel and six reported). This includes six missense mutations (CRYAA-R12C, R21W, R54C, CRYAB- A171T, CRYGC-R168W, CRYGS- S39C), two nonsense mutations (CRYBB2- Q155X, CRYGD- R140X), and one splice mutation, which was identified in two families (CRYBA1-IVS3+1G>A).
Conclusions
Crystallin mutations are responsible for 16.6% of the inherited pediatric cataract in this population. As causative mutations have not been found in many of the families analyzed, this study suggests the presence of further novel genes or sequence elements involved in the pathogenesis of cataract in these families.
Introduction
Pediatric cataracts are common and represent one of the most treatable causes of lifelong visual impairment. It is estimated that globally, 20 million children under the age of 16 suffer from cataract, and among these, 200,000 (15%) are severely visually impaired or blind [1,2]. While this figure is relatively low compared to the 17 million (40%) adults who are blind due to cataract [3], the burden of disability in terms of “blind-years” is huge due to the child’s life expectancy after developing the visual disability. In addition, untreated congenital cataracts can cause permanent blindness by interfering with the focus of light on the retina, necessary for formation of neural pathways necessary for vision. This presents an enormous problem in developing countries in terms of human morbidity, economic loss, and social burden.
Pediatric cataracts are both clinically and genetically heterogeneous. About one-third to one-half of all bilateral pediatric cataracts have a genetic basis [4-6]. All three forms of Mendelian inheritance have been observed, and the most frequently seen in non-consanguineous populations is autosomal dominant (AD) transmission [7,8]. To date, at least 34 loci in the human genome have been reported to be associated with various forms of pediatric cataract. Of the mapped loci, mutations have been identified in 22 specific genes including encoding crystallins (CRYAA [9], CRYAB [10], CRYBA1 [11], CRYBA4 [12], CRYBB1 [13], CRYBB2 [14], CRYBB3 [15], CRYGC, CRYGD [16], and CRYGS [17]), cytoskeletal proteins (BFSP1 [18] and BFSP2 [19]), membrane proteins (GJA3 [20] and GJA8 [21], MIP [22] and LIM2 [23]), transcription factors (HSF4 [24], PITX3 [25], and MAF [26]), glucosaminyl (N-acetyl) transferase 2 (GCNT2 [27]), chromatin modifying protein-4B (CHMP4B [28]), and TMEM114 [29]. On the basis of current studies, mutations in about half of affected families occur in crystallins, a quarter in connexins, and the remainder is evenly split between membrane proteins, intermediate filament proteins, and transcription factors. However, the relative contribution of these classes of genes to pediatric cataracts in India is still unclear.
Crystallins are the major cytoplasmic proteins of the lens and their stability and appropriate interactions are critical for lens transparency. Crystallin genes encode more than 95% of the water soluble structural proteins present in the vertebrate lens and their encoded proteins account for more than 30% of its mass. In 1894, Morner first separated bovine lens proteins into three soluble fractions and one insoluble fraction [30]. The soluble fractions consisted of α-, β-, and γ-crystallins, which are found in all vertebrate lenses and are referred to as “ubiquitous crystallins.” In the mature human lens, α-crystallin makes up roughly 40%, β-crystallin 35%, and γ-crystallin 25% of the total crystallin protein. The β- and γ-crystallins form a super family as they share a common two domain structure composed of four extremely stable, torqued β-pleated sheets termed “Greek key” motifs. At least 13 functional crystallin genes have been identified in humans, and of these, 10 major crystallin genes have been associated with pediatric cataract [12,31].
To clarify the relative contributions of mutations in the crystallin genes to congenital cataracts in the Indian population, a systematic screening of the 10 crystallin genes associated with cataract was performed in a large panel of congenital cataract patients.
Methods
Family ascertainment
Patients with a positive family history of childhood cataract were recruited from the Pediatric Ophthalmology Clinic, Aravind Eye Hospital (AEH), Madurai, India. The family members were interviewed to obtain a detailed medical, ophthalmic, and family history and were included in the study based on their availability and willingness. Patients with a history suggestive of intrauterine infection such as rubella, complicated cataract, and traumatic cataract were excluded from the study.
Ophthalmologic examination included the best corrected visual acuity (Snellen’s), slit lamp biomicroscopy, intraocular pressure measurement by applanation tonometry, and fundus examination. Corneal diameter and axial length were measured when indicated. In addition, the probands were examined by a physician to check for the presence of other systemic abnormalities. Photographs of significant findings were taken depending upon the cooperation of the patient. Normal subjects were drawn from the General Ophthalmology Clinic of the Aravind Eye Hospital (AEH) matching the ethnic distribution of the pediatric cataract patients. Ethics approval for the study was obtained from the Institutional Review Boards of the AEH and Combined Neuroscience IRB at the National Eye Institute, and the study was performed in accordance to the tenets of Declaration of Helsinki. Informed consent was obtained from the participating members in the study. DNA was extracted from venous blood as previously described [32].
Molecular analysis
One affected representative individual from each of the identified families was chosen for mutation analysis. Primers were designed to amplify the entire coding exons and 10–30 bp of the flanking intronic sequences of the 10 crystallin genes - CRYAA, CRYAB, CRYBA1, CRYBA4, CRYBB1, CRYBB2, CRYBB3, CRYGC, CRYGD, and CRYGS (primer sequences available upon request). Single strand conformational polymorphism (SSCP) analysis was employed in the detection of mutations. The amplicons were denatured at 98 °C for 5 min and electrophoresed at 700–800 V for 8 −12 h at room temperature on 6%–12% polyacrylamide gels. Additives including glycerol (5%–10%) were used in gels. The gels were silver stained according to the modified protocol of Bassam et al. [33].The samples whose electrophoresis patterns differed from those of the controls were sequenced. The polymerase chain reaction (PCR) product was purified using Ampure and CleanSeq purification kits (Agencourt, Beckman, Beverly, MA) on a Beckman NX MC automated workstation (Beckman). Sequencing of the DNA was performed using chain terminator chemistry with a Big Dye terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) on an ABI 3130 DNA analysis system.
The DNA sequence was analyzed and compared with the reference sequence using the Seqman program of the DNASTAR analysis package (Lasergene, Madison, WI). If a nonsynonymous sequence change that results in a change in the amino acid sequence of the protein was found, available family members were then analyzed for cosegregation of the genotype with the phenotype. In addition, 100 control chromosomes were analyzed to confirm association of a previously described mutation, and 200 normal control chromosomes were analyzed to confirm the association of a novel sequence variation with the disease phenotype. If available, the presence or the absence of a restriction site was employed to confirm the cosegregation of sequence variation among the family members and in the control population. When no suitable restriction site was identified, direct sequencing was performed to analyze the cosegregation of the genotype with the disease phenotype.
Homology modeling
Modeling of the human βγ-crystallins, γC- and γS-crystallin, was performed by homology modeling based on correspondent crystal coordinates for murine protein structures (Brookhaven Protein Database [PDB] files: 2v2u and 1zw0) as the structural templates [34]. For each model, primary sequences were aligned by the method of Needleman and Wunsch [35] and incorporated in the program, Look version 3.5.2 [36,37] for three-dimensional structure prediction. Finally, two models of γC- and γS-crystallin monomers were built by the automatic segment matching method in the program, Look, followed by 500 cycles of non-bound energy minimization [38]. These models were used to predict the effect of mutations in CRYGC and CRYGS. In addition, structures of two human βγ-crystallins, γD- and βB2-crystallin, (PDB files: 1ytq and 1hk0, respectively) were chosen to model mutations identified in CRYGD and CRYBB2. The conformations of the proteins with mutations Q155X (βB2-crystallin), R168W (γC-crystallin), R140X (γD-crystallin), and S39C (γS-crystallin) were modeled and refined by self-consistent ensemble optimization (500 cycles) [37].
Results and Discussion
A total of 60 patients below the age of 25 years with a positive family history of pediatric cataract were registered for the study. The families were coded as CCW1-CCW60 according to the order collected. The mode of inheritance was inferred from the pedigree based on the vertical or horizontal transmission as dominant or recessive inheritance, respectively. About 53% (n=32) had AD inheritance, 30% (n=18) had autosomal recessive (AR) inheritance, and in 17% (n=10), the inheritance pattern could not be determined. The age of onset was recorded as the age at which the disease was first noticed by the child’s parents or first documented by a clinician. Depending on the age of onset, the pediatric cataract was classified as congenital, infantile, or juvenile cataract. Congenital cataracts present at birth were observed in 56% (n=33) of the patient families. An additional 8% (n=5) had infantile cataracts that developed within the first three years of life, and cataracts that developed within the second decade of life and were characterized as juvenile cataract were observed in 33% (n=20). Finally, 3% of the families (n=2) had a variable age of onset. Fifteen families also revealed other associated ocular disorders such as microcornea, microphthalmia, myopia, and anterior and/or posterior lenticonus.
The mutations identified in the crystallin genes analyzed are summarized in Table 1. The analysis allowed the identification of causative nine mutations in 10 of the 60 families. Of the nine individual mutations identified, three are novel and six have been reported previously. These include six missense mutations (CRYAA-R12C, R21W, R54C, CRYAB-A171T, CRYGC-R168W, and CRYGS-S39C), two nonsense mutations (CRYBB2-Q155X, CRYGD-R140X), and one splice mutation that was identified in two separate families (CRYBA1-IVS3+1G>A). This suggests that mutations in crystallin genes might account for approximately 16.6% of pediatric cataracts in the study population.
Table 1. Summary of crystallin mutations identified in south Indian families.
|
Gene |
Family ID |
Base change |
Amino acid |
Blosum 80 |
Age of onset |
Cataract phenotype |
Other ocular anomalies |
Restriction site employed in conformation |
Controls screened |
|
CRYAA |
CCW-46 |
c.104 C>T |
R12C |
−6 |
Birth |
Nuclear |
Microcornea |
ApaLI + |
100 |
| CCW-36 |
c.130 C>T |
R21W |
−5 |
Birth |
Nuclear |
Microcornea |
MspI - |
100 |
|
| CCW-55 |
c.230 C>T |
R54C |
−6 |
Birth |
Nuclear |
Microcornea + Microphthalmous |
HpyCH4V + |
100 |
|
|
CRYAB |
CCW-22 |
c.557G>A |
A171T* |
0 |
Birth |
Lamellar |
- |
HpyF10VI - |
100 |
|
CRYBA1 |
CCW-01 |
IVs3+1G>A |
- |
- |
Birth |
Lamellar, Floriform (Variable) |
- |
Nla III + |
100 |
| CCW-57 |
IVs+1G>A |
- |
- |
Birth |
Lamellar |
- |
|||
|
CRYBB2 |
CCW-19 |
c.495C>T |
Q155X |
−8 |
5–10 years |
Cortical + pulverulent |
- |
- |
50 |
|
CRRYGC |
CCW-33 |
c.502 C>T |
R168W |
−5 |
Birth |
Lamellar |
- |
- |
50 |
|
CRYGD |
CCW-45 |
c.418C>T |
R140X* |
−8 |
Birth |
Nuclear |
- |
- |
100 |
| CRYGS | CCW-47 | c.168C>G | S39C* | −2 | 7–10 years | Lamellar, Sutural (Variable) | - | HpyF10VI + | 100 |
The asterisk indicates the novel mutations identified in this study. The “+” and “−” indicate the gain and loss of a restriction site as a result of the mutation.
Summary of mutations in α-crystallin genes and the corresponding proteins
The α-crystallin gene family consists of two similar genes coding for αA-crystallin (CRYAA located on chromosome 21q22.3) and αB-crystallin (CRYAB on chromosome 11q22.1) sharing 57% sequence identity [39]. The first exon of each gene encodes 60 amino acids consisting of a repeated 30 amino acid motif while the second and third exons code for regions homologous to the small heat shock proteins [39,40].
Description of mutations in CRYAA, their inheritance and associated phenotypes
In the present study, mutation analysis of CRYAA revealed three missense mutations, c.104 C>T (R12C) in family CCW46, c.130 C>T (R21W) in family CCW36, and c.230 C>T (R54C) in family CCW55 (Figure 1A-C). It is interesting to note that the affected members of these three families developed bilateral congenital nuclear cataract in association with microcornea except for family CCW36 (R21W) where the affected members were also diagnosed with microphthalmia. All three mutations identified in this study occur outside the small heat shock protein core domain (sHSP), and the arginines at positions 12, 21, and 54 are highly conserved among α-crystallin (Figure 2). Previous studies on the predicted topology of CRYAA suggest that the NH2-terminal region is involved in quaternary subunit interactions [41], raising the possibility that inappropriate disulfide bridge formation by the cysteine in the R12C and R54C mutations might lead to insolubility of the protein. Another possibility is that αA-crystallin has been shown to have a strong tendency to maintain a constant net charge through evolution [42]. The mutations identified in this study result in the replacement of the basic amino acid, arginine, by a neutral one, which also could affect protein–protein interactions.
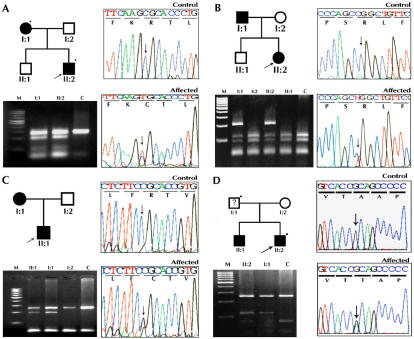
Figure 1.
Pedigree, electropherogram, and restriction fragment length polymorphism of ADCC families with a mutation in the α-crystallin gene. In the pedigree, the square symbol represents males while the circle symbol represents females. A circle with a slash denotes a deceased individual, and a blackened symbol denotes an affected individual. The dot on the upper right corner of the symbol means a sample was available from that individual, and the arrow denotes the proband. A, B, C show mutation analyses of CRYAA. A: Family CCW46 shows a heterozygous c.104 C>T resulting in a novel ApaLI restriction site (mutant allele−191 and 63 bp, wild type−254 bp) B: Family CCW36 shows a c.130 C>T change that results in the loss of the MspI restriction site (mutant allele-254 bp, wild type-116, 90, and 48 bp) C: Family CCW55 shows the gain of a novel HpyCH4V site cosegregating with the affected individual heterozygous for c.230 C>T transition (mutant allele-254, 191, and 63 bp and wild type-254 bp). D: Family CCW22 has a mutation in CRYAB, c.557G>A. This mutation results in the loss of a HpyF10VI site (mutant allele-258 and 118 bp, wild type-258, 77, and 41 bp). M denotes 100 bp DNA ladder, and C denotes unrelated control.
Figure 2.
Protein sequence alignments. Protein sequence alignment showing sequence conservation of CRYAA (top alignment), CRYAB (second alignment), CRYGC (third alignment), and CRYGD (bottom alignment) among closely related species and members of the α- and γ-crystallin gene families.
An increasing number of CRYAA mutations have been associated with cataract formation in humans and mice, exhibiting both autosomal dominant and recessive inheritance. Interestingly, most of the mutations in αA-crystallin involve arginine residues and act in a dominant fashion (R12C [42], R21W/L [43,44], R49C [45], G98R [46], and R116C/H [9,43]). These mutations have led to isolated cataracts or cataracts associated with microcornea, microphthalmia, and/or iris coloboma [9,43,44]. The changes, R12C and R21W, have previously been reported to result in the cataract-microcornea syndrome [43]. The cataract phenotypes of these two mutations are similar, consisting of central, zonular opacification with varying involvement of the anterior and posterior pole. The identical mutations identified in this study result in nuclear cataracts with microcornea. The CRYAA mutation, W9X, in humans and the R54H mutation in mice have been reported to cause AR cataract [47,48]. During the preparation of this manuscript, Khan et al. [49] reported R54C to cause an autosomal recessive cataract where the family members homozygous for the mutation had congenital total cataract with microcornea and bilateral punctuate cataracts were observed in heterozygous carriers of the mutation. In contrast, the R54C mutation reported in this study is associated with autosomal dominant congenital nuclear cataract (ADCC) associated with microcornea. In summary, the CRYAA mutations identified in this study result in nuclear cataract in association with microcornea in all three families and associated with additional microphthalmia in one family.
Description of mutations in CRYAB, their inheritance and associated phenotypes
A novel mutation, A171T, was identified in CRYAB in family CCW22 (Figure 1D), the proband was the only available clinically confirmed affected family member available for this study. The proband and his unexamined father revealed a heterozygous c.557G>A change. The proband showed lamellar cataract phenotype with no ocular or systemic abnormalities.
αB-crystallin is widely expressed in several non-ocular tissues including cardiac and skeletal muscles [50]. Several mutations in CRYAB have been described including one associated with cataract and desmin related myopathy (Arg120Gly) [51] and in patients with desmin-related myofibrillar myopathy without cataract (464delCT and G151X) [52]. In 2001, the CRYAB mutation, K150fs, was associated with isolated posterior polar cataract in an English family [10]. P20S and D140N have also been reported to cause isolated forms of ADCC [53], but it is not clear why some mutations in CRYAB cause muscle system disorders and/or cardiovascular defects and other mutation causes isolated cataract.
The A171T mutation identified in this study could not be confirmed to cosegregate with cataracts due to the lack of available family members. However, analysis of 200 normal control chromosomes showed the absence of such a change. This mutation also lies in a phylogenetically conserved region in the COOH-terminii of αB-crystallin (Figure 2), which is considered necessary for the chaperone function of the protein. Most of the mutations reported in CRYAB are in this region, consistent with the importance of the sequence encoded in exon 3 for αB-crystallin function. Further, there have been reports of a CRYAB mutation associated with lamellar cataract [53] similar to the phenotype observed in the proband of family CCW22. While not definitive, all of these suggest that the A171T mutation is responsible for the cataracts in this patient.
Summary of mutations in β-crystallin genes and the corresponding proteins
The β-crystallins are divided into seven subgroups, three basic (βB1-, βB2-, and βB3-crystallin) located on chromosome 22q11.2 and four acidic forms (βA1/βA3-crystallin located on chromosome 17q11.2, βA2-crystallin located on chromosome 2q33, and βA4-crystallin located on chromosome 22q11.2). All the β-crystallin genes except CRYBA2 have been associated with congenital cataracts. Screening β-crystallin genes in this population revealed a splice site mutation in CRYBA1 and a nonsense mutation in CRYBB2.
Description of mutations in CRYBA1, their inheritance and associated phenotypes
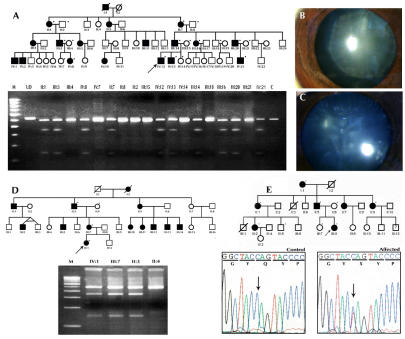
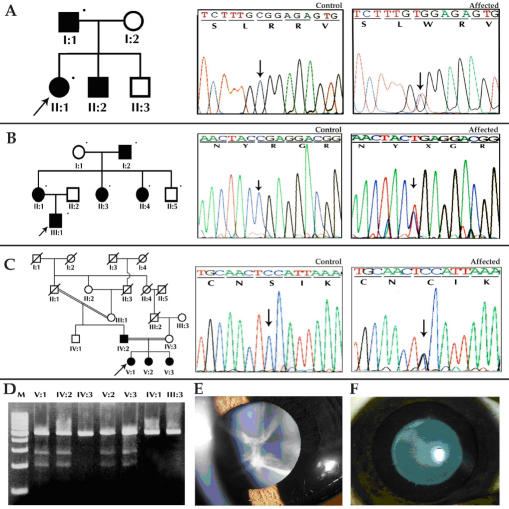
A previously reported splice site mutation (IVS3+1 G>A) was identified in two families, CCW1 and CCW57, with an AD mode of inheritance (Figure 3A,D). The disease shows complete penetrance, and the ophthalmic records confirm the opacification of the lens was bilateral in all affected members. The members whose medical records were available in these two families showed the affected individuals were diagnosed with zonular lamellar opacification. However, in family CCW1, individual IV:21 had lamellar cataract and needed to undergo surgery at two years of age while his father III:20 at 33 years old had floriform cataract and his visual acuity is 6/36 in the right eye and 4/60 in the left (Figure 3B,C).
Figure 3.
Mutation analysis of β-crystallin genes. A: Pedigree and RFLP analysis of family CCW1 is shown. The dot on the upper right corner of the symbol denotes a sample was available for the study. The mutation, IVS3+1 G>A, in CRYBA1 creates a novel NlaIII restriction site. Wild type individuals display a 488 bp and 32 bp band while the affected individual display a 488 bp, 346 bp, 142 bp, and 32 bp cleavage product. The latter 32 bp DNA fragment expected to be generated from the PCR product due to the presence of a common NlaIII site cannot be visualized in the agarose gel. Lens images of individuals of family CCW-1, IV:12 aged 8 years with lamellar cataract (B) and individual III:20 aged 33 years (C), show floriform cataract respectively. D: Pedigree and RFLP analysis of family CCW58 with IVS3+1 G>A mutation in CRYBA1 are shown. E: Mutation analysis of CRYBB2 in pedigree CCW19 shows heterozygous C>T transition in the chromatogram that results in Q155X. M denotes 100 bp DNA ladder, C denotes unrelated control, and UD denotes undigested PCR product.
To date, there are three mutations in CRYBA1 that have been associated with cataracts. These include a three base pair deletion in exon 4 (∆G91) that caused AD congenital pulverulent nuclear cataract in three Chinese families, a suture sparing nuclear cataract in a Swiss family and an English family with the identical mutation for which no clinical description is available [54-57]. A splice site mutation, IVS3+1G>C, has been associated with a pulverulent cataract phenotype in a family from Brazil [58], and a G>A transition at the same position has been reported to cause lamellar, sutural cataract in an Indian family and sutural nuclear cataract with peripheral cortical opacities in an Australian pedigree [11,59]. The CRYBA1 mutation identified in the two families (CCW1 and CCW57) in this study is a G>A transition at the 5′ donor splice site. The mutation was found to be associated with a characteristic zonular lamellar cataract. The Australian pedigree with the IVS3+1G>A mutation included an individual with a mild opacity. Similar individuals are included in our family (CCW1). The cause of intra-pedigree variability of cataract phenotype is unknown, although it could result as an effect of a modifier gene or environmental factor.
The donor splice site mutation is expected to cause disruption of βA3- and βA1-crystallins, which differ from each other only in the length of their 5′-terminal extensions. The first two exons of CRYBA1 encode the NH2-terminal arm, and exons 3–6 encode the Greek key motifs [60]. As speculated by Kannabiran et al. [11], the effect of this mutation will result in skipping of the donor splice junction or recruitment of a cryptic splice site that would affect the proper formation of the Greek key motifs.
Description of mutations in CRYBB2, their inheritance and associated phenotypes
As previously reported, the nonsense mutation, Q155X (c.495C>T), in exon 6 of CRYBB2 was identified in family CCW19 (Figure 3E). Affected individuals in this three-generation family have cortical cataracts with pulverulent opacities. All affected individuals developed poor vision between the ages five and seven years. The mutation (Q155X) is predicted to remove the final 51 amino acids, effectively deleting ~90% of the fourth Greek key motif and the entire COOH-terminal arm of the protein as demonstrated in Figure 4A. The loss of these amino acids is predicted to remove the last three β-strands in the sequence, resulting in an unstable molecule.
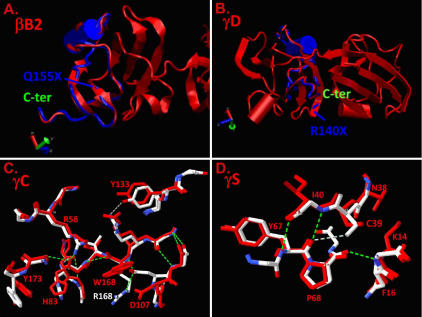
Figure 4.
Molecular modeling of the effects of mutations presented in Table 1 on βγ-crystallin structures. Normal structures are shown in red and the overlaid mutant structures are shown in blue (truncations) and blue and white (substitutions). A: The normal βB2-crystallin structure is shown in red, and the Q155X mutation is predicted to remove the COOH-terminal 51 amino acids shown in the blue overlay. B: The normal γD-crystallin structure is shown in red and the R140X mutation is predicted to remove the COOH-terminal 34 amino acids shown in the blue overlay. C: The normal γC-crystallin structure in the vicinity of the R168R mutation is shown in white with positive charges shown in blue and negative charges in red. The overlaid R168W mutant is shown in red, demonstrating the substitution of the polar arginine residue by the apolar tryptophan and consequent displacement of D107. D: The normal γS-crystallin structure in the vicinity of the fragments of 3D structures of mutant proteins (γC-crystallin) and S39C (γS-crystallin) mutations are shown in red. Hydrogen-bonding patterns of the mutant γC- and γS-crystallin structures are shown by dashed lines (green).
Previously, six geographically distinct families have been reported with the same chain-termination mutation (Q155X) in CRYBB2 [14,61-65]. Clinical descriptions of the cataract morphologies vary widely from cerulean cataract in American and Moroccan families [14,61] to a Coppock-like phenotype with pulverulent opacities in a Swiss family [62], punctate cataracts with cerulean opacification in an Indian family [63], and progressive polymorphic cataracts ranging from pulverulent to dot, strip, star-like, and sheet shapes in a Chinese family [64]. A Chilean family exhibited variable and disparate phenotypes ranging from pulverulent cortical opacities to a dense posterior star-shaped subcapsular cataract [65]. The cataract morphology identified in this study shows little similarity to the Swiss and Chilean families. This mutation clearly demonstrates the phenotypic heterogeneity of the disease.
Summary of mutations in γ-crystallin genes and the corresponding proteins
The family of γ-crystallin genes is mainly located in a cluster of six highly related genes (CRYGA-CRYGF) on human chromosome 2q33–35 and the seventh CRYG gene (CRYGS) on human chromosome 3. Mutations in CRYGC, CRYGD, and CRYGS have been associated with congenital and juvenile hereditary cataracts [16,17,66]. Molecular analysis of these genes in this study revealed one previously reported mutation in CRYGC (R168W) and novel mutations in CRYGD (R140X) and CRYGS (S39C). Results of modeling the structural changes corresponding to these mutations are shown in Figure 4B-D.
Description of mutations in CRYGC, their inheritance and associated phenotypes
Two affected members of family CCW33, both having an R168W mutation, were available for study (Figure 5A). Hospital records confirmed the cataract was present at birth. Upon clinical examination, affected individual II:1 displayed a lamellar cataract phenotype (Figure 5F). R168 is highly conserved among the γ-crystallins (Figure 2). It is a hydrophilic amino acid with a positive charge and lies within the extended strand on the surface of the molecule, interacting with water. In the R168W mutant, arginine is replaced by tryptophan, a hydrophobic amino acid. Changing the solvation property of an amino acid residue on the surface of the γ-crystallin protein is predicted to diminish its solubility [67]. In addition, replacement of R168 with a tryptophan residue is predicted to result in a significant change in the conformation of residues located close to residue 168, altering the hydrogen bonding pattern as demonstrated in Figure 4C.
Figure 5.
Mutation analysis of γ-crystallin genes. A: Family CCW-33 shows an R168W mutation in CRYGC. B: Family CCW-45 shows an R140X mutation in CRYGD. C: Family CCW47 shows a S39C mutation in CRYGS. D: RFLP analysis of CRYGS exon 1 shows the loss of HpyF10VI site (mutant allele-274 bp and 204 bp, wild-type allele-478 bp). E: Individual V:3 of family CCW47 shows sutural cataract. F: Individual II:1 of family CCW33 shows lamellar cataract. M denotes 100 bp DNA ladder, and C denotes unrelated control.
To date, there are three mutations reported in CRYGC, one insertion mutation and two missense mutations. The insertion mutation (p.C42fs) resulted in zonular, perinuclear, or polymorphic cataracts with varying degrees of opacification [68]. Of the missense mutations, T5P is reported to cause a Coppock-like cataract that presents with a dust-like opacity of the fetal nucleus [16] and R168W has been reported to cause lamellar and dense nuclear cataracts in Indian and Mexican families, respectively [69,70]. It is interesting to note that the lamellar cataract morphology observed in family CCW33 in this study is similar to that reported by Santhiya et al. [69] in that both are childhood lamellar cataracts. While both these families are from southern India, a single origin could not be confirmed.
Description of mutations in CRYGD, their inheritance and associated phenotypes
A nonsense mutation in exon 3 of CRYGD (c.418C>T, R140X) cosegregates with cataracts in the affected members of family CCW45 (Figure 5B). Ophthalmic records of the proband confirm the opacity was present during early infancy and mainly affected the central zone or the fetal nucleus. All other affected members of the family underwent surgery at an early age. The R140X mutation is predicted to result in the loss of 34 amino acids, resulting in only partial formation of fourth Greek key motif and misfolding of the truncated protein. The truncated part of the R140X mutant is shown in blue (Figure 4B). This would presumably be followed by precipitation of the protein resulting in opacification of the lens. The two previously reported nonsense mutations in CRYGD, Y134X in Danish families [42] and W157X reported in an Indian family [69], also occur in exon 3, affecting the fourth Greek key motif and causing nuclear cataract. The R140X mutation identified in this study results in a similar nuclear cataract phenotype.
Description of mutations in CRYGS, their inheritance and associated phenotypes
A novel missense mutation (c.168C>G, S39C) was identified in exon 2 of CRYGS in the two-generation family CCW47 (Figure 5C). Affected individuals in this family show a progressive juvenile onset cataract. Deterioration of vision of affected family members began at seven years of age. Initially, one eye had more severe opacification than the other. The sequence variation, c.168C>G, in CRYGS cosegregates with the trait in the kindred studied and is absent in 200 ethnically matched normal control chromosomes (Figure 5D). Serine is highly conserved at position 39 (Figure 2). Replacement of serine at position 39 by cysteine as shown in Figure 4D is predicted to break three hydrogen bonds including the one involving S39 directly (shown in white). Although both serine and cysteine are neutral hydrophilic amino acids, it is possible that the new cysteine residue might form inappropriate homocysteine bonds in an oxidizing environment.
This is the second report of a CRYGS mutation to be associated with cataract. In the previous report, a G18V mutation was identified in a Chinese family affected with progressive polymorphic cortical cataract. The opacities in this Chinese family varied not only between family members but also between eyes in the same patient [17]. Similar findings were observed in family CCW47 in which the severity of the cataracts varied from one eye to another of the same patient. Individual V:3 shows sutural opacities in the left eye while the right eye showed no opacification (Figure 5E). Individual V:1 and V:2 had lamellar opacities and underwent surgery at the end of the first decade of their lives. Phenotypic variation in the size and density of the opacities and in their position was also observed among other affected family members.
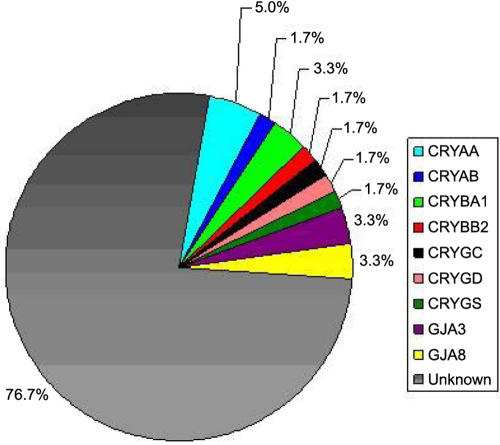
In summary, this study identified mutations in 10 of 60 families from southern India affected by pediatric cataracts. CRYAA was the gene most commonly involved with causative mutations identified in 5% of the families while both CRYAA and CRYAB together accounted for 6.6% of pediatric cataracts in the families studied. Identical mutations were identified in CRYBA1 (IVS3+1G>A) in two large families, suggesting that this is a common mutation site for this gene. No mutations were detected in CRYBA4, CRYBB1, or CRYBB3. It is notable that all of the mutations identified in this study cause autosomal dominant cataract. Previously, the same set of families has been screened for mutations in connexin genes (GJA3 [71] and GJA8 [72]), which revealed mutations in four families. The frequency of involvement of the 10 crystallin genes and two connexin genes analyzed in the Indian families is summarized in Figure 6. Taken together, these 12 genes account for almost one-quarter of inherited cataracts in this group of families from southern India.
Figure 6.
Frequency of crystallin and connexin mutations in the south Indian population. Pie chart showing the frequencies of crystallin and connexin mutations in the south Indian popultaion as seen in this study and in references [71] and [72].
In contrast, causative mutations in these 12 genes (10 crystallin and two connexin) were not identified in over 76% of the families (n=46). While it is certainly possible that causative mutations occurring in these genes were missed in some cases or would not be detected by sequencing (e.g., large deletions or mutations in promoter or intronic splice enhancer sequences), it seems unlikely that these would account for a large fraction of the families without identified mutations. Thus, the pathogenesis of the majority of cataract families in India seems not to be accounted for mutations by the coding exons of crystallins or connexins, and mutations in lens crystallins and connexins appear to be underrepresented in this population. A previous Australian study involving systematic screening for mutation in five crystallin genes (CRYAA, CRYBA1, CRYBB2, CRYGC, and CRYGD) in 38 pedigrees has reported only two mutations (CRYBA1-IVS3+1G>A and CRYGD-P23T). Given that studies on two independent populations describe a low frequency of crystallin mutations, perhaps the dominance of crystallins in the literature is not a reflection of the true distribution of cataract genes, but rather it represents the attractiveness of the crystallins as candidate genes for screening studies. However the possibility cannot be excluded that mutations in one of the other reported candidate genes such as membrane intrinsic proteins (MIP and LIM2), intermediate filament protein (BFSP2), transcription factors (MAF, PITX3, and HSF4), and glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (GCNT2) may be the major cause of hereditary cataracts in India. Alternatively, it is possible that currently unidentified genes may be a more significant cause of cataracts than previously thought. Extensive linkage analysis and screening of additional candidate genes will assist in discrimination between the above hypotheses.
Acknowledgments
We thank all the members of the participating family. Thanks are also due to B. Suganthalakshmi, A. Gomathy, and Jeya Krishnan for their technical assistance. This study was supported by Aravind Eye Hospital (Madurai, TN, India) and National Eye Institute (NIH, Bethesda, MD).
References
- 1.Johnson, GJ, editor. The epidemiology of eye disease. 2nd ed. London: Arnold; 2003. [Google Scholar]
- 2.Foster A, Gilbert C, Rahi J. Epidemiology of cataract in childhood: a global perspective. J Cataract Refract Surg. 1997;23:601–4. doi: 10.1016/s0886-3350(97)80040-5. [DOI] [PubMed] [Google Scholar]
- 3.Dawson CR, Schwab IR. Epidemiology of cataract-a major cause of preventable blindness. Bull World Health Organ. 1981;59:493–501. [PMC free article] [PubMed] [Google Scholar]
- 4.Francois J. Genetics of cataract. Ophthalmologica. 1982;184:61–71. doi: 10.1159/000309186. [DOI] [PubMed] [Google Scholar]
- 5.Merin S, Crawford JS. The etiology of congenital cataracts: A survey of 386 cases. Can J Ophthalmol. 1971;6:178–82. [PubMed] [Google Scholar]
- 6.Haargaard B, Wohlfahrt J, Fledelius HC, Rosenberg T, Melbye M. A nationwide Danish study of 1027 cases of congenital/infantile cataracts: etiological and clinical classifications. Ophthalmology. 2004;111:2292–8. doi: 10.1016/j.ophtha.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 7.Francis PJ, Berry V, Bhattacharya SS, Moore AT. The genetics of childhood cataract. J Med Genet. 2000;37:481–8. doi: 10.1136/jmg.37.7.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Francis PJ, Berry V, Hardcastle AJ, Maher ER, Moore AT, Bhattacharya SS. A locus for isolated cataract on human Xp. J Med Genet. 2002;39:105–9. doi: 10.1136/jmg.39.2.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Litt M, Kramer P, LaMorticella DM, Murphey W, Lovrien EW, Weleber RG. Autosomal dominant congenital cataract associated with a missense mutation in the human alpha crystallin gene CRYAA. Hum Mol Genet. 1998;7:471–4. doi: 10.1093/hmg/7.3.471. [DOI] [PubMed] [Google Scholar]
- 10.Berry V, Francis P, Reddy MA, Collyer D, Vithana E, MacKay I, Dawson G, Carey AH, Moore A, Bhattacharya SS, Quinlan RA. Alpha-B crystallin gene (CRYAB) mutation causes dominant congenital posterior polar cataract in humans. Am J Hum Genet. 2001;69:1141–5. doi: 10.1086/324158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kannabiran C, Rogan PK, Olmos L, Basti S, Rao GN, Kaiser-Kupfer M, Hejtmancik JF. Autosomal dominant zonular cataract with sutural opacities is associated with a splice mutation in the betaA3/A1-crystallin gene. Mol Vis. 1998;4:21. [PubMed] [Google Scholar]
- 12.Billingsley G, Santhiya ST, Paterson AD, Ogata K, Wodak S, Hosseini SM, Manisastry SM, Vijayalakshmi P, Gopinath PM, Graw J, Heon E. CRYBA4, a novel human cataract gene, is also involved in microphthalmia. Am J Hum Genet. 2006;79:702–9. doi: 10.1086/507712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mackay DS, Boskovska OB, Knopf HL, Lampi KJ, Shiels A. A nonsense mutation in CRYBB1 associated with autosomal dominant cataract linked to human chromosome 22q. Am J Hum Genet. 2002;71:1216–21. doi: 10.1086/344212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Litt M, Valenzuela CR, Lamorticella DM, Schultz DW, Mitchell TN, Kramer P, Maumenee IH. Autosomal dominant cerulean cataract is associated with a chain termination mutation in the human β-crystallin gene CRYBB2. Hum Mol Genet. 1997;6:665–8. doi: 10.1093/hmg/6.5.665. [DOI] [PubMed] [Google Scholar]
- 15.Riazuddin SA, Yasmeen A, Yao W, Sergeev YV, Zhang Q, Zulfiqar F, Riaz A, Riazuddin S, Hejtmancik FJ. Mutations in βB-crystallin associated with autosomal recessive cataract in two Pakistani families. Invest Ophthalmol Vis Sci. 2005;46:2100–6. doi: 10.1167/iovs.04-1481. [DOI] [PubMed] [Google Scholar]
- 16.Heon E, Priston M, Schorderet DF, Billingsley GD, Girad PO, Lubsen N, Munier FL. The γ-Crystallins and humans cataracts: A puzzle made clearer. Am J Hum Genet. 1999;65:1261–7. doi: 10.1086/302619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun H, Ma Z, Li Y, Liu B, Li Z, Ding X, Gao Y, Ma W, Tang X, Li X, Shen Y. Gamma-S crystallin gene (CRYGS) mutation causes dominant progressive cortical cataract in humans. J Med Genet. 2005;42:706–10. doi: 10.1136/jmg.2004.028274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramachandran RD, Perumalsamy V, Hejtmancik JF. Autosomal recessive juvenile onset cataract associated with mutation in BFSP1. Hum Genet. 2007;121:475–82. doi: 10.1007/s00439-006-0319-6. [DOI] [PubMed] [Google Scholar]
- 19.Conley YP, Erturk D, Keverline A, Mah TS, Keravala A, Barnes LR, Bruchis A, Hess JF, Fitzgerald PG, Weeks DE, Ferrell RE, Gorin MB. A juvenile-onset, progressive cataract locus on chromosome 3q21-q22 is associated with a missense mutation in the beaded filament structural protein-2. Am J Hum Genet. 2000;66:1426–31. doi: 10.1086/302871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rees MI, Watts P, Fenton I, Clarke A, Snell RG, Owen MJ, Gray J. Further evidence of autosomal dominant congenital zonular pulverulent cataracts linked to 13q11 (CZP3) and a novel mutation in connexin 46 (GJA3). Hum Genet. 2000;106:206–9. doi: 10.1007/s004390051029. [DOI] [PubMed] [Google Scholar]
- 21.Shiels A, Mackay D, Ionides A, Berry V, Moore A, Bhattacharya S. A missense mutation in the human connexin50 gene (GJA8) underlies autosomal dominant “zonular pulverulent” cataract, on chromosome 1q. Am J Hum Genet. 1998;62:526–32. doi: 10.1086/301762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berry V, Francis P, Kaushal S, Moore A, Bhattacharya S. Missense mutations in MIP underlie autosomal dominant “polymorphic” and lamellar cataracts linked to 12q. Nat Genet. 2000;25:15–7. doi: 10.1038/75538. [DOI] [PubMed] [Google Scholar]
- 23.Pras E, Levy-Nissenbaum E, Bakhan T, Lahat H, Assia E, Geffen-Carmi N, Frydman M, Goldman B, Pras E. A missense mutation in the LIM2 gene is associated with autosomal recessive presenile cataract in an inbred Iraqi Jewish family. Am J Hum Genet. 2002;70:1363–7. doi: 10.1086/340318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bu L, Jin Y, Shi Y, Chu R, Ban A, Eiberg H, Andres L, Jiang H, Zheng G, Qian M, Cui B, Xia Y, Liu J, Hu L, Zhao G, Hayden MR, Kong X. Mutant DNA-binding domain of HSF4 is associated with autosomal dominant lamellar and Marner cataract. Nat Genet. 2002;31:276–8. doi: 10.1038/ng921. [DOI] [PubMed] [Google Scholar]
- 25.Semina EV, Ferrell RE, Mintz-Hittner HA, Bitoun P, Alward WL, Reiter RS, Funkhauser C, Daack-Hirsch S, Murray JC. A novel homeobox gene PITX3 is mutated in families with autosomal-dominant cataracts and ASMD. Nat Genet. 1998;19:167–70. doi: 10.1038/527. [DOI] [PubMed] [Google Scholar]
- 26.Vanita V, Singh D, Robinson PN, Sperling K, Singh JR. A novel mutation in the DNA-binding domain of MAF at 16q23.1 associated with autosomal dominant “cerulean cataract” in an Indian family. Am J Med Genet A. 2006;140:558–66. doi: 10.1002/ajmg.a.31126. [DOI] [PubMed] [Google Scholar]
- 27.Pras E, Raz J, Yahalom V, Frydman M, Garzozi HJ, Pras E, Hejtmancik JF. A nonsense mutation in the glucosaminyl (N-acetyl) transferase 2 gene (GCNT2): association with autosomal recessive congenital cataracts. Invest Ophthalmol Vis Sci. 2004;45:1940–5. doi: 10.1167/iovs.03-1117. [DOI] [PubMed] [Google Scholar]
- 28.Shiels A, Bennett TM, Knopf HL, Yamada K, Yoshiura K, Niikawa N, Shim S, Hanson PI. CHMP4B, a novel gene for autosomal dominant cataracts linked to chromosome 20q. Am J Hum Genet. 2007;81:596–606. doi: 10.1086/519980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jamieson RV, Farrar N, Stewart K, Perveen R, Mihelec M, Carette M, Grigg JR, McAvoy JW, Lovicu FJ, Tam PP, Scambler P, Lloyd IC, Donnai D, Black GC. Characterization of a familial t(16;22) balanced translocation associated with congenital cataract leads to identification of a novel gene, TMEM114, expressed in the lens and disrupted by the translocation. Hum Mutat. 2007;28:968–77. doi: 10.1002/humu.20545. [DOI] [PubMed] [Google Scholar]
- 30.Morner CT. Untersuchungender protein-substanzen in den lichtbrechenden Medien des Auges. Hoppe Seylers Z Physiol Chem. 1894;18:61–106.. [Google Scholar]
- 31.Shiels A, Hejtmancik JF. Genetic origins of cataract. Arch Ophthalmol. 2007;125:165–73. doi: 10.1001/archopht.125.2.165. [DOI] [PubMed] [Google Scholar]
- 32.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bassam BJ, Caetano-Anolles G, Gresshoff PM.Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 199119680–3..Erratum inAnal Biochem 1991198 217 [DOI] [PubMed] [Google Scholar]
- 34.Abola E, Bernstein FC, Bryant SH, Koetzle TF, Weng J. Protein data bank. In: Allen FH, Bergerhoff G, Sievers R, editors. Crystallographic databases-information content, software systems, scientific applications. Cambridge: Data Commission of the International Union of Crystallography; 1987. p.107–32. [Google Scholar]
- 35.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–53. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 36.Lee C, Subbiah S. Prediction of protein side-chain conformation by packing optimization. J Mol Biol. 1991;217:373–88. doi: 10.1016/0022-2836(91)90550-p. [DOI] [PubMed] [Google Scholar]
- 37.Lee C. Predicting protein mutant energetics by self-consistent ensemble optimization. J Mol Biol. 1994;236:918–39. doi: 10.1006/jmbi.1994.1198. [DOI] [PubMed] [Google Scholar]
- 38.Levitt M. Accurate modeling of protein conformation by automatic segment matching. J Mol Biol. 1992;226:507–33. doi: 10.1016/0022-2836(92)90964-l. [DOI] [PubMed] [Google Scholar]
- 39.de Jong WW, Terwindt EC, Bloemendal H. The amino acid sequence of the A chain of human alpha-crystallin. FEBS Lett. 1975;58:310–3. doi: 10.1016/0014-5793(75)80286-9. [DOI] [PubMed] [Google Scholar]
- 40.Wistow G. Domain structure and evolution in alpha-crystallins and small heat-shock proteins. FEBS Lett. 1985;181:1–6. doi: 10.1016/0014-5793(85)81102-9. [DOI] [PubMed] [Google Scholar]
- 41.Koteiche HA, Mchaourab HS. Folding pattern of the alpha crystallin domain in alpha A-crystallin determined by site directed spin labeling. J Mol Biol. 1999;294:561–77. doi: 10.1006/jmbi.1999.3242. [DOI] [PubMed] [Google Scholar]
- 42.Berengian AR, Bova MP, Mchaourab HS. Structure and function of the conserved domain in alphaA-crystallin. Site-directed spin labeling identifies a beta-strand located near a subunit interface. Biochemistry. 1997;36:9951–7. doi: 10.1021/bi9712347. [DOI] [PubMed] [Google Scholar]
- 43.Hansen L, Yao W, Eiberg H, Kjaer KW, Baggesen K, Hejtmancik JF, Rosenberg T. Genetic heterogeneity in microcornea-cataract: five novel mutations in CRYAA, CRYGD, and GJA8. Invest Ophthalmol Vis Sci. 2007;48:3937–44. doi: 10.1167/iovs.07-0013. [DOI] [PubMed] [Google Scholar]
- 44.Graw J, Klopp N, Illig T, Preising MN, Lorenz B. Congenital cataract and macular hypoplasia in humans associated with a de novo mutation in CRYAA and compound heterozygous mutations in P. Graefes Arch Clin Exp Ophthalmol. 2006;244:912–9. doi: 10.1007/s00417-005-0234-x. [DOI] [PubMed] [Google Scholar]
- 45.Mackay DS, Andley UP, Shiels A. Cell death triggered by a novel mutation in the alphaA-crystallin gene underlies autosomal dominant cataract linked to chromosome 21q. Eur J Hum Genet. 2003;11:784–93. doi: 10.1038/sj.ejhg.5201046. [DOI] [PubMed] [Google Scholar]
- 46.Santhiya ST, Soker T, Klopp N, Illig T, Prakash MV, Selvaraj B, Gopinath PM, Graw J. Identification of a novel, putative cataract-causing allele in CRYAA (G98R) in an Indian family. Mol Vis. 2006;12:768–73. [PubMed] [Google Scholar]
- 47.Pras E, Frydman M, Levy-Nissenbaum E, Bakhan T, Raz J, Assia EI, Goldman B, Pras E. A nonsense mutation (W9X) in CRYAA causes autosomal recessive cataract in an inbred Jewish Persian family. Invest Ophthalmol Vis Sci. 2000;41:3511–5. [PubMed] [Google Scholar]
- 48.Chang B, Hawes NL, Roderick TH, Smith RS, Eckenlively JR, Horwitz J. And Davisson MT. Identification of a missense mutation in the αA-crystallin gene of the lop18 mouse. Mol Vis. 1999;5:21. [PubMed] [Google Scholar]
- 49.Khan AO, Aldahmesh MA, Meyer B. Recessive Congenital Total Cataract with Microcornea and Heterozygote Carrier Signs Caused by a Novel Missense CRYAA Mutation (R54C). Am J Ophthalmol. 2007;144:949–52. doi: 10.1016/j.ajo.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 50.Dubin RA, Wawrousek EF, Piatigorsky J. Expression of the murine alpha B-crystallin gene is not restricted to the lens. Mol Cell Biol. 1989;9:1083–91. doi: 10.1128/mcb.9.3.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vicart P, Caron A, Guicheney P, Li Z, Prevost MC, Faure A, Chateau D, Chapon F, Tome F, Dupret JM, Paulin D, Fardeau M. A missense mutation in the αB-crystallin chaperone gene causes a desmin-related myopathy. Nat Genet. 1998;20:92–5. doi: 10.1038/1765. [DOI] [PubMed] [Google Scholar]
- 52.Selcen D, Engel AG. Myofibrillar myopathy caused by novel dominant negative alpha B-crystallin mutations. Ann Neurol. 2003;54:804–10. doi: 10.1002/ana.10767. [DOI] [PubMed] [Google Scholar]
- 53.Liu M, Ke T, Wang Z, Yang Q, Chang W, Jiang F, Tang Z, Li H, Ren X, Wang X, Wang T, Li Q, Yang J, Liu J, Wang QK. Identification of a CRYAB mutation associated with autosomal dominant posterior polar cataract in a Chinese family. Invest Ophthalmol Vis Sci. 2006;47:3461–6. doi: 10.1167/iovs.05-1438. [DOI] [PubMed] [Google Scholar]
- 54.Qi Y, Jia H, Huang S, Lin H, Gu J, Su H, Zhang T, Gao Y, Qu L, Li D, Li Y. A deletion mutation in the βa1/a3 crystallin gene (cryba1/a3) is associated with autosomal dominant congenital nuclear cataract in a chinese family. Hum Genet. 2004;114:192–7. doi: 10.1007/s00439-003-1049-7. [DOI] [PubMed] [Google Scholar]
- 55.Ferrini W, Schorderet DF, Girarad OP, Uffer S, Heon E, Munier FL. CRYBA3/A1 gene mutation associated with suture-sparing autosomal dominant congenital nuclear cataract: A novel phenotype. Invest Ophthalmol Vis Sci. 2004;45:1436–41. doi: 10.1167/iovs.03-0760. [DOI] [PubMed] [Google Scholar]
- 56.Reddy MA, Bateman OA, Chakarova C, Ferris J, Berry V, Lomas E, Sarra R, Smith MA, Moore AT, Bhattacharya SS, Slingsby C. Characterization of the G91del CRYBA1/3-crystallin protein: a cause of human inherited cataract. Hum Mol Genet. 2004;13:945–53. doi: 10.1093/hmg/ddh110. [DOI] [PubMed] [Google Scholar]
- 57.Lu S, Zhao C, Jiao H, Kere J, Tang X, Zhao F, Zhang X, Zhao K, Larsson C. Two Chinese families with pulverulent congenital cataracts and ΔG91 CRYBA1 mutations. Mol Vis. 2007;13:1154–60. [PubMed] [Google Scholar]
- 58.Bateman JB, Geyer DD, Flodman P, Johannes M, Sikela J, Walter N, Moreira AT, Clancy K, Spence MA. A new betaA1-crystallin splice junction mutation in autosomal dominant cataract. Invest Ophthalmol Vis Sci. 2000;41:3278–85. [PubMed] [Google Scholar]
- 59.Burdon KP, Wirth MG, Mackay DA, Russell-Eggitt IM, Craig JE, Elder JE, Dickinson JL, Sale MM. Investigation of crystallingenes in familial cataract, and report of two disease associated mutations. Br J Ophthalmol. 2004;88:79–83. doi: 10.1136/bjo.88.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hogg D, Tsui LC, Gorin M, Breitman ML. Characterization of the human beta-crystallin gene Hu beta A3/A1 reveals ancestral relationships among the beta gamma-crystallin superfamily. J Biol Chem. 1986;261:12420–7. [PubMed] [Google Scholar]
- 61.Hilal L, Nandrot E, Belmekki M, Chefchaouni M, El Bacha S, Benazzouz B, Hajaji Y, Gribouval O, Dufier J, Abitbol M, Berraho A. Evidence of clinical and genetic heterogeneity in autosomal dominant congenital cerulean cataracts. Ophthalmic Genet. 2002;23:199–208. doi: 10.1076/opge.23.4.199.13881. [DOI] [PubMed] [Google Scholar]
- 62.Gill D, Klose R, Munier FL, McFadden M, Priston M, Billingsley G, Ducrey N, Schorderet DF, Heon E. Genetic heterogeneity of the Coppock-like cataract: a mutation in CRYBB2 on chromosome 22q11.2. Invest Ophthalmol Vis Sci. 2000;41:159–65. [PubMed] [Google Scholar]
- 63.Vanita , Sarhadi V, Reis A, Jung M, Singh D, Sperling K, Singh JR, Bürger J. A unique form of autosomal dominant cataract explained by gene conversion between betacrystallin B2 and its pseudogene. J Med Genet. 2001;38:392–6. doi: 10.1136/jmg.38.6.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yao K, Tang X, Shentu X, Wang K, Rao H, Xia K. Progressive polymorphic congenital cataract caused by a CRYBB2 mutation in a Chinese family. Mol Vis. 2005;11:758–63. [PubMed] [Google Scholar]
- 65.Bateman JB, von-Bischhoffshaunsen FR, Richter L, Flodman P, Burch D, Spence MA. Gene conversion mutation in crystallin, beta-B2 (CRYBB2) in a Chilean family with autosomal dominant cataract. Ophthalmology. 2007;114:425–32. doi: 10.1016/j.ophtha.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 66.Stephan DA, Gillanders E, Vanderveen D, Freasletz D, Wistow G, Baxevanis AD, Robbins CM, Van Auken A, Quesenberry MI, Bailey-Wilson J, Juo SH, Trent JM, Smith L, Brownstein MJ. Progressive juvenile-onset punctuate cataracts caused by mutation of the γD-crystallin gene. Proc Natl Acad Sci USA. 1999;96:1008–12. doi: 10.1073/pnas.96.3.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Evans P, Wyatt K, Wistow GJ, Bateman OA, Wallace BA, Slingsby C. The P23T cataract mutation causes loss of solubility of folded gammaD-crystallin. J Mol Biol. 2004;343:435–44. doi: 10.1016/j.jmb.2004.08.050. [DOI] [PubMed] [Google Scholar]
- 68.Ren Z, Li A, Shastry BS, Padma T, Ayyagari R, Scott MH, Parks MM, Kaiser-Kupfer MI, Hejtmancik JF. A 5-base insertion in the gammaC-crystallin gene is associated with autosomal dominant variable zonular pulverulent cataract. Hum Genet. 2000;106:531–7. doi: 10.1007/s004390000289. [DOI] [PubMed] [Google Scholar]
- 69.Santhiya ST, Manohar SM, Rawlley D, Vijayalakshmi P, Namperumalsamy P, Gopinath PM, Loster J, Graw J. Novel mutations in the γ-crystallin genes caused autosomal dominant congenital cataracts. J Med Genet. 2002;39:352–8. doi: 10.1136/jmg.39.5.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gonzalez-Huerta LM, Messina-Baas OM, Cuevas-Covarrubias SA. A family with autosomal dominant primary congenital cataract associated with a CRYGC mutation: evidence of clinical heterogeneity. Mol Vis. 2007;13:1333–8. [PubMed] [Google Scholar]
- 71.Devi RR, Reena C, Vijayalakshmi P. Novel mutations in GJA3 associated with autosomal dominant congenital cataract in the Indian population. Mol Vis. 2005;11:846–52. [PubMed] [Google Scholar]
- 72.Devi RR, Vijayalakshmi P. Novel mutations in GJA8 associated with autosomal dominant congenital cataract and microcornea. Mol Vis. 2006;12:190–5. [PubMed] [Google Scholar]