Figure 5. Disproportionate over-representation of general transcription factors and chromatin remodeling genes in undifferentiated ES cells.
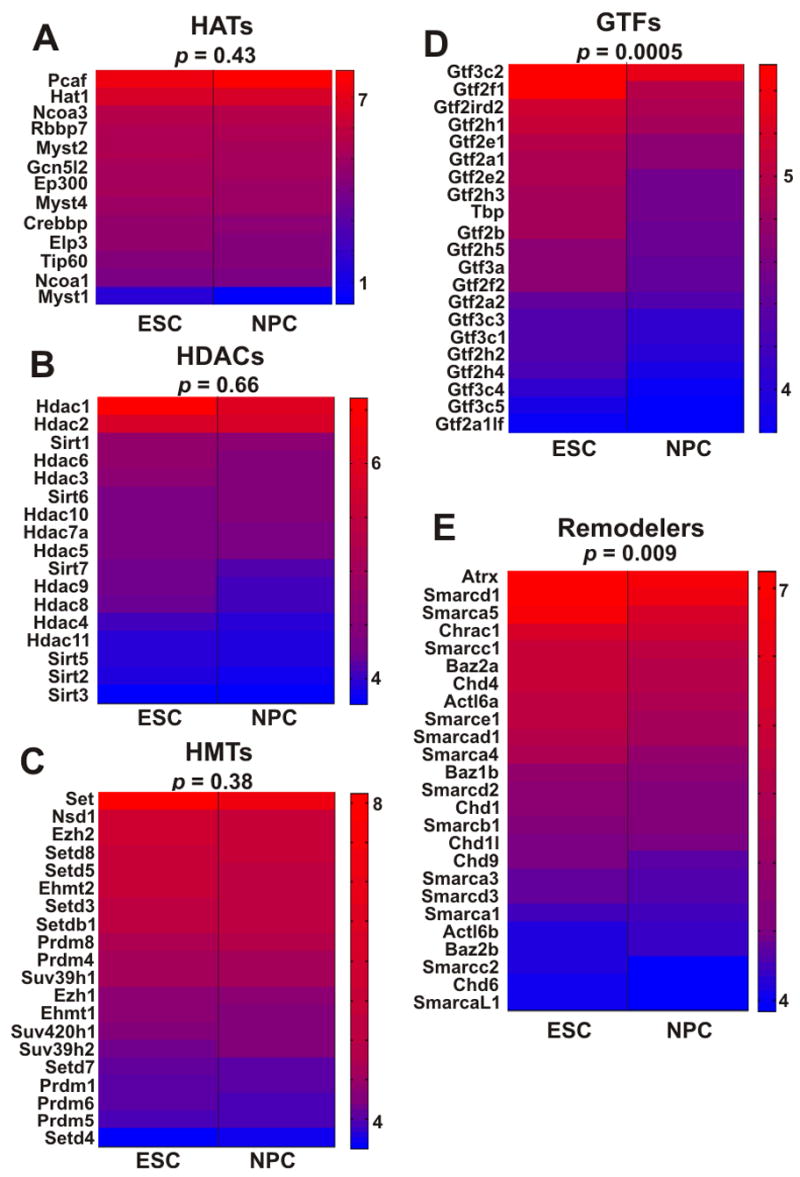
(A–E) Transcription level heat-maps of different groups of genes that are associated with transcription and regulation of chromatin, including histone acetyltransferases (A), histone deacetylases (B), histone methyltransferases (C), general transcription factors (D) and chromatin remodeling proteins (E). Gene names are given on the left of each map, p values (binomial hypothesis testing) are indicated on top. Chromatin remodeling factors and general transcription factors are disproportionately expressed in ES cells. Heat-maps were generated using microarray signal levels displayed as arbitrary units. Red-to-blue corresponds to high-to-low signal intensity.
