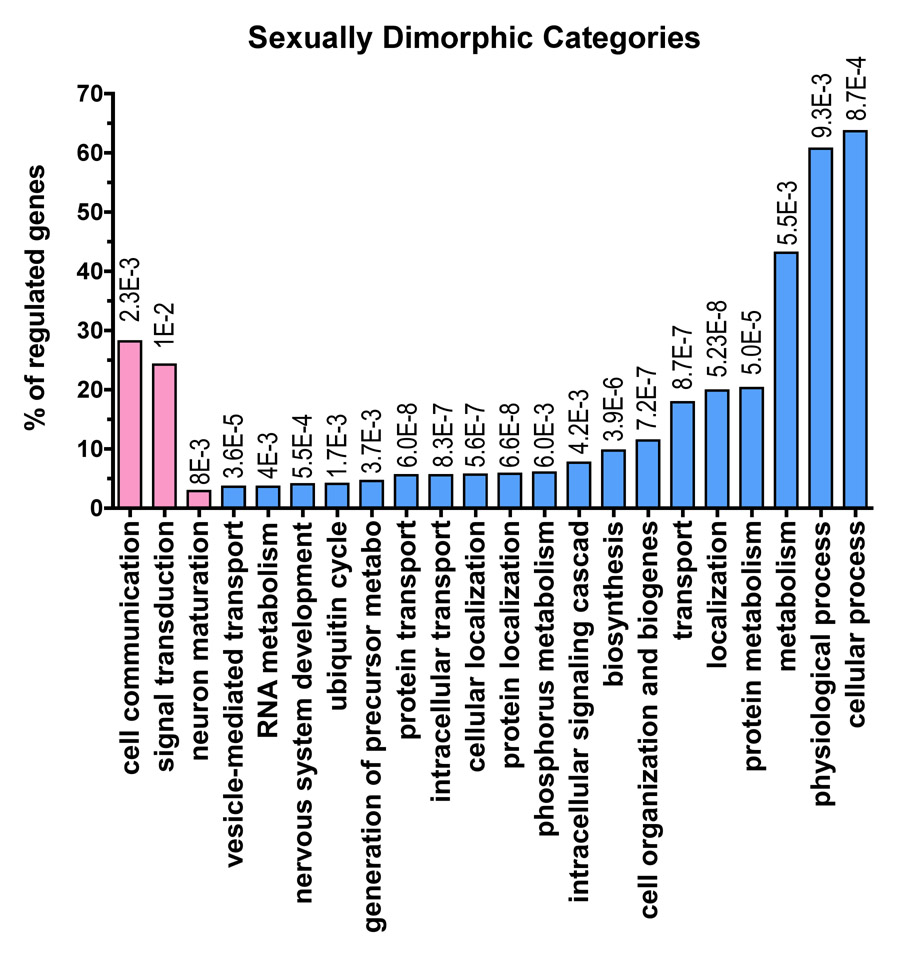
Figure 2. Functional profiling of the gender-specific genes.
124 probe-sets (~ 120 genes) upregulated in the females relative to males, and over 2060 probe-sets (~2000 genes) upregulated in the males relative to females were assigned to functional categories using the Gene Ontology Database. For each functional grouping, the probability that the number of genes observed was greater than chance was calculated according to the hypergeometric distribution (see methods). Only the probe-sets with a multiclass q value<0.11 were used for functional analysis (1870 probe-sets). The table shows the categories identified as significant (p <0.01, # genes in category ≥3% of total regulated genes) for the biological process: Pk= p value; k/u= number of regulated genes observed (k) over the total number of genes in the GO class (u). The regulated genes can be represented under more than one functional term as they can be implicated in one or more biological processes. A list of the genes regulated by gender can be found as supplementary Table 1. The functional categories overrepresented in the gene sets upregulated in the females are shown with the pink bars and the functional categories overrepresented in the gene sets upregulated in males are shown with the light blue bars. At the top of each bar is shown the p value for each functional category. In this figure we show all of the functional categories that are significantly different according to our method. We show both more specific and informative categories such as ‘ubiquitin cycle’ and more broad categories such as ‘cellular process’. Cellular process in only one of eighteen different sub-categories under the parent term ‘biological process’. Some of the categories at the same hierarchical level as ‘cellular process’ are ‘biological adhesion’, ‘reproductive process’ or ‘response to stimulus’ (to search categories and their meaning and hierarchical relationship see QuickGO: http://www.ebi.ac.uk/ego/).