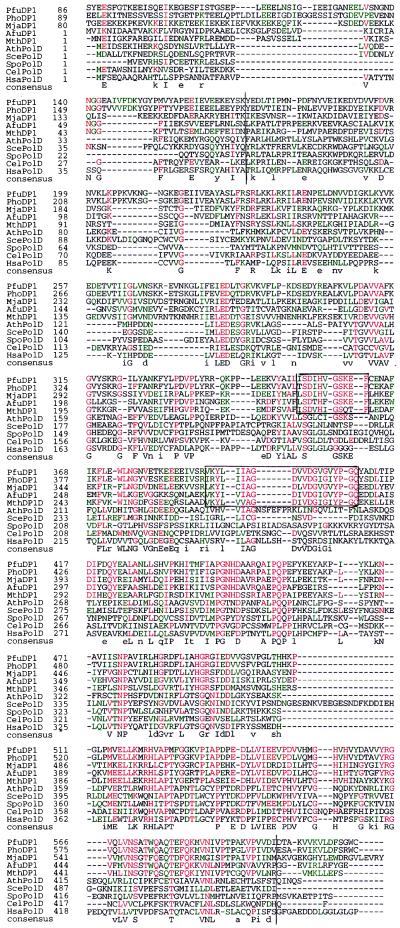
Figure 1.
Amino acid sequence alignment of the euryarchaeal DP1 and eukaryotic Pol δ small subunit. The sequences were aligned with clustalw, version 1.7. The polymerases shown are from P. furiosus (Pfu), P. horikoshii (Pho), M. jannaschii (Mja), A. fulgidus (Afu), M. thermoautotrophicum (Mth), A. thaliana (Ath), S. cerevisiae (Sce), S. pombe (Spo), C. elegans (Cel), and H. sapiens (Hsa). The ORF names of the archaeal proteins obtained from their genome projects were Mja DP1 (MJ0702), Afu DP1 (AF1790), Mth DP1 (MTH1405), and Pho DP1 (PHBN023). Amino acid residues that are identical (red) or similar (green) in ≥50% of the positions are indicated. Upper case letters indicate consensus residues with identities at ≥50% positions, including, at least, one from each domain. Lowercase letters indicate the most frequent residues including, at least, one from each domain. The highly conserved regions from the eury archaeal group are boxed. Amino acids with similar properties are grouped as LIMV, AG, YWF, DEQN, KRH, and ST.