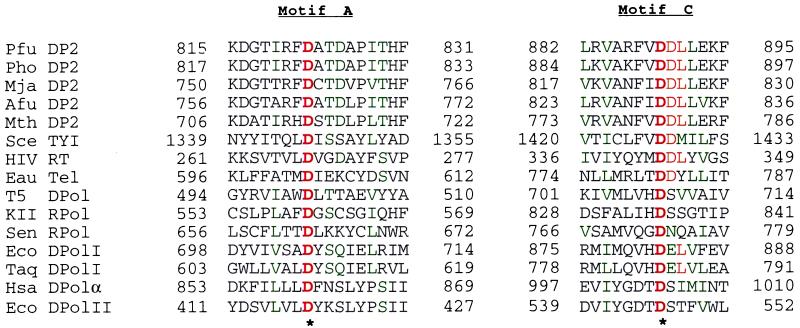
Figure 2.
Amino acid sequence alignment showing two major conserved regions of polymerases, including the DP2 of Euryarchaeota. The sequences were aligned with clustalw, version 1.7. The asterisks indicate invariant residues. Amino acid residues that are identical (red) or similar (green) in ≥50% of the positions are indicated. The polymerases shown are from P. furiosus (Pfu), P. horikoshii (Pho), M. jannaschii (Mja), A. fulgidus (Afu), M. thermoautotrophicum (Mth), S. cerevisiae transposon TYI protein B (Sce TYI), HIV DNA polymerase polyprotein (HIV RT), E. aediculatus telomerase subunit (Eau Tel), bacteriophage T5 DNA polymerase (T5 DPol), bacteriophage KII RNA polymerase (KII Rpol), Sendai virus RNA polymerase β subunit (Sen Rpol), E. coli DNA polymerase I (Eco DPolI), T. aquaticus Pol I (Taq DPolI), H. sapiens DNA polymerase α (Hsa), and E. coli Pol II (Eco DpolII). The ORF names of the archaeal proteins obtained from their genome projects are Mja DP2 (MJ1630), Afu DP2 (AF1722), Mth DP2 (MTH1536), and Pho DP2 (PHBN021). Amino acids were classified as in Fig. 1.