Abstract
Folded proteins can exist in multiple conformational substates. Each substate reflects a local minimum on the free-energy landscape with a distinct structure. By using ultrafast 2D-IR vibrational echo chemical-exchange spectroscopy, conformational switching between two well defined substates of a myoglobin mutant is observed on the ≈50-ps time scale. The conformational dynamics are directly measured through the growth of cross peaks in the 2D-IR spectra of CO bound to the heme active site. The conformational switching involves motion of the distal histidine/E helix that changes the location of the imidazole side group of the histidine. The exchange between substates changes the frequency of the CO, which is detected by the time dependence of the 2D-IR vibrational echo spectrum. These results demonstrate that interconversion between protein conformational substates can occur on very fast time scales. The implications for larger structural changes that occur on much longer time scales are discussed.
Keywords: multidimensional IR spectroscopy, myoglobin, protein dynamics, protein structural change, ultrafast IR
A folded protein with a particular structure occupies a minimum on its free-energy landscape (1–3). However, the minimum is frequently a local minimum. Other minima of similar energy can also exist. When a protein occupies any one of these minima, it has a distinct structure. The different structures are substates of the folded protein. Transitions from one minimum to another correspond to dynamical changes in the structure of the protein that take the protein from one substate to another. Under thermal equilibrium conditions, there will be continual conformational switching among substates. In addition to interconversion between substates, proteins undergo continuous structural fluctuations within a particular substate minimum. Such fluctuations within a substate minimum give rise to processes such as small ligand “diffusion” through a protein to an active site (4).
The ability of proteins to undergo conformational switching is central to protein function. When an enzyme binds a substrate, the protein conformation will change (5). On the path of protein folding, a protein will sample many conformations as it progresses toward the native folded structure (6). Proteins can undergo large global conformational changes, which occur on long time scales, milliseconds to seconds. However, these large, slow conformational changes, such as those that occur after substrate binding to an enzyme, involve a vast number of more local elementary conformational steps.
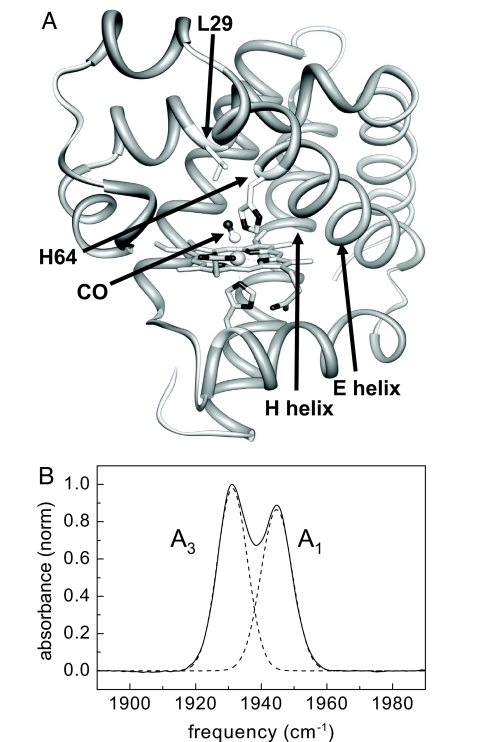
The experimental determination for the time scale of elementary conformational steps is a long-standing problem that has now been addressed successfully by using ultrafast 2D-IR vibrational echo chemical-exchange spectroscopy. The problem of substate switching has been studied extensively for the protein myoglobin (Mb) with the ligand CO bound at the active site (MbCO) (7–9). The Fourier transform IR (FT-IR) spectrum of the heme-ligated CO stretching mode of Mb has three absorption bands, denoted A0 (1,965 cm−1), A1 (1,945 cm−1), and A3 (1,932 cm−1) (10). MbCO interconverts among these three conformational substates under thermal equilibrium. The distal histidine, His-64, has a prominent role in determining the conformational substates of Mb (Fig. 1A). Changes in the configuration of the distal histidine cause its imidazole side group to move relative to the CO (10, 11). When the conformation of the distal histidine results in the imidazole being rotated out of the heme pocket, the interaction of the distal histidine with the ligated CO becomes weak. This out-of-the-pocket configuration gives rise to the A0 band (9, 12). The distal histidine conformation that places the imidazole in the heme pocket gives rise to two substates, A1 and A3, which involve strong interactions of the imidazole side group with the CO ligand. The lower frequency of A3 compared with A1 reflects a closer proximity of the imidazole to the CO in A3 (11, 13, 14). Each A substate exhibits a distinct ligand binding rate (2, 7). Therefore, the three peaks in the FT-IR spectrum of MbCO reflect functionally distinct conformational substates.
Fig. 1.
Structure of CO-bound myoglobin. (A) Crystal structure of MbCO taken from the Protein Data Bank (ID code 1MWC). The distal histidine, His-64; the CO; the location of Leu-29; and the E helix are indicated. (B) FT-IR spectrum of the CO stretch of heme-ligated CO for the myoglobin mutant L29I (solid curve). The spectrum was fitted by two Gaussians (dashed curves), which represent the absorption bands of the A1 and A3 substates.
By using flash photolysis measurements, the kinetic rate constant for the A0 − A1/A3 conformational switching, in which the distal histidine swings its imidazole side group out of and into the heme pocket, is estimated to be in the range of ≈1–10 μs (8, 13). These indirect measurements based on ligand binding rate constants have also been applied to the A1 and A3 interconversion by using low-temperature flash photolysis studies. The results were extrapolated to ambient temperature and yield <1 ns for switching between A1 and A3 (13). Molecular dynamics (MD) simulations have placed the A1–A3 switching time on the order of 100–200 ps (14).
It has been difficult to directly measure fast elementary substate interconversion times, including the A1-A3 switching. Here, we report a direct measurement of the A1–A3 substate switching time under thermal equilibrium conditions by using 2D-IR vibrational echo chemical-exchange spectroscopy. This method has recently proven useful for studying fast dynamical processes in liquids (15–17). The 2D-IR vibrational echo chemical-exchange experiment is akin to a 2D NMR chemical-exchange experiment, except that it can operate on a picosecond time scale, and it directly probes the structural degrees of freedom through the time evolution of the 2D vibrational spectrum. The A1 and A3 peaks in the FT-IR spectrum appear as bands on the diagonal of the 2D-IR vibrational echo spectrum. As time increases, cross peaks grow in because of the interconversion between the A1 and A3 substates (chemical exchange). Analysis of the time-dependent growth of the chemical-exchange peaks yields the A1 going to A3 interconversion time of 47 ps.
Results and Discussion
The FT-IR spectrum of wild-type MbCO shows three discrete CO stretching bands, the A0, A1, and A3 bands (7, 10, 18). The A1 substate of wild-type MbCO is predominately populated and dominates the absorption spectrum. Because the A3 absorption peak of wild-type MbCO is relatively small compared with the A1 peak, we performed the experiments on an Mb mutant, L29I, in which a leucine is replaced with an isoleucine (10). This small change causes the A1 and A3 bands in the FT-IR spectrum to be approximately the same size (Fig. 1B). Like wild-type MbCO, the amount of A0 conformation is small in the L29I mutant (≈5%). Changes at position 29 cause a slight modification of the configuration of the distal histidine because the amino acid residue at position 29 interacts with the imidazole side chain of the distal histidine (Fig. 1A) (10). In the background-subtracted FT-IR spectrum shown in Fig. 1B, the two L29I CO absorption bands are at 1,932 cm−1 and 1,945 cm−1. These bands correspond to the A3 and A1 substates, respectively (10). The peak positions for the CO absorption bands of L29I are essentially identical to those of wild-type Mb within the reported significant figures (10).
Experimental details for performing 2D-IR vibrational echoes and chemical-exchange experiments have been presented previously (15, 17, 19, 20). Qualitatively, the experimental method works in the following manner. Three ultrashort IR pulses tuned to the frequency of the A1 and A3 vibrational modes are crossed in the sample. The IR excitation pulses, 110 fs and 150 cm−1 FWHM, are produced with a Ti:sapphire regenerative amplifier-pumped optical parametric oscillator. Because the pulses are very short, they have a broad bandwidth that makes it possible to simultaneously excite the vibrational modes of both the A1 and A3 substates. The first laser pulse “labels” the initial structures of the species by defining their initial frequency, ωτ. The second pulse ends the first time period τ and starts clocking the reaction time period Tw during which the labeled species undergo substate switching (chemical exchange) and vibrational relaxation to the ground state. The third pulse ends the population dynamics period of length Tw and begins a third period of length ≤τ, which ends with the emission of the vibrational echo pulse of frequency ωm. The vibrational echo signal reads out information about the final structures of all labeled species by their frequency ωm.
There are two types of time periods in the experiment. The times between pulses 1 and 2 and between pulse 3 and the vibrational echo pulse are called coherence periods. During these periods, the vibrations are in coherent superpositions of two vibrational states. Fast vibrational oscillator frequency fluctuations induced by fast structural fluctuations of the protein cause dynamic dephasing, which is one contribution to the line shapes in the conventional 1D absorption spectrum. During the period Tw between pulses 2 and 3, called the population period, a vibration is in a vibrational eigenstate. Slower structural fluctuations of the protein cause the CO frequency to evolve in time. This time evolution of the frequency, termed spectral diffusion, contributes to the 2D line shapes. Other processes during the population period, particularly chemical exchange, also produce changes in the 2D spectrum. Chemical exchange occurs when two species in equilibrium interconvert without changing the overall number of either species. Interconversion back and forth between the A1 and A3 substates is a type of chemical exchange. In other contexts, it has been demonstrated that chemical exchange causes new cross peaks to grow in as Tw is increased (15–17, 21–23). The growth of the cross peaks with increasing time is directly related to the chemical-exchange rate (15–17, 21–23). Here, the growth of the cross peaks in the 2D vibrational echo spectrum of L29I with increasing Tw is used to extract the A1–A3 substate interconversion rate.
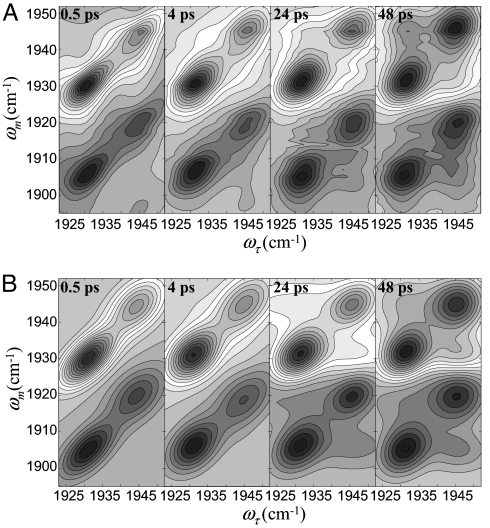
Fig. 2A shows 2D-IR spectra of CO bound to L29I at several Tw values. The bands in the upper half of the spectrum are positive-going and correspond to the 0–1 vibrational transitions. The bands in the lower half of the spectrum are negative-going, which arise from vibrational echo emission at the frequency of the 1–2 vibrational transition. The negative-going bands are shifted along the ωm axis by the anharmonicity of CO stretching mode (24, 25). The data have been normalized to the largest peak at each Tw. Consider the spectrum for Tw = 0.5 ps. The two bands on the diagonal correspond to the two peaks in the absorption spectrum shown in Fig. 1B. These two bands are centered at (ωτ, ωm) = (1,932 cm−1, 1,932 cm−1) and (1,945 cm−1, 1,945 cm−1) corresponding to the A3 and A1 substates, respectively. The off-diagonal band centered at (ωτ, ωm) = (1,932 cm−1, 1,908 cm−1) and (1,945 cm−1, 1,922 cm−1) are from vibrational echo emission at the 1–2 vibrational transitions of the A3 and A1 substates, respectively. At Tw = 0.5 ps, there are no cross peaks in the 0–1 and 1–2 regions because 0.5 ps is short relative to the conformational switching time.
Fig. 2.
2D-IR spectra of CO bound to L29I. (A) 2D-IR spectra of L29I-CO at various times, Tw. Each contour corresponds to a 10% signal change. The bands in the upper half of the spectrum (positive-going) correspond to the 0–1 vibrational transition. The bands in the lower half of the spectrum (negative-going) arise from vibrational echo emission at the 1–2 transition frequency. (B) Calculated 2D-IR spectra of L29I-CO at various times, Tw, using the known input parameters and the substate switching time constant obtained from fitting the data.
In contrast, by Tw = 48 ps (Fig. 2A) enough time has elapsed for conformational switching to occur to a significant extent. The conformational switching is manifested by the growth of cross peaks (17), which are most apparent in the upper left portion of the 0–1 bands and the lower right portion of the 1–2 bands. These cross peaks correspond to the A3 switching to A1 and A1 switching to A3, respectively. Because the system is in equilibrium, the rate of A1 converting into A3 is equal to the rate of A3 converting into A1. If the anharmonicity is sufficiently large, so that the positive-going 0–1 bands do not overlap with the negative-going 1–2 bands, then the cross peaks appear symmetrically about the 0–1 diagonal peaks and the corresponding 1–2 peaks (17). Here, the overlap of the positive- and negative-going bands reduces the amplitudes of the other two cross peaks, that is, the 0–1 A1 to A3 cross peak at (ωτ, ωm) = (1,945 cm−1, 1,932 cm−1) and the 1–2 A3 to A1 cross peak at (ωτ, ωm) = (1,932 cm−1, 1,920 cm−1).
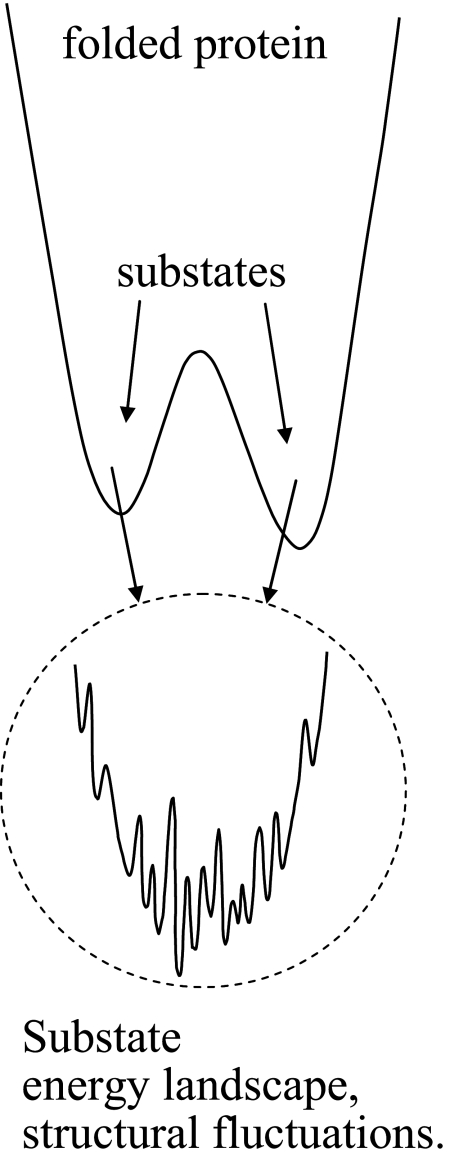
In addition to the growth of the cross peaks in Fig. 2A, the shapes of the bands change with increasing Tw. At short Tw, the 2D line shapes show significant inhomogeneous broadening. Inhomogeneous broadening is evidenced by elongation along the diagonal. As time proceeds, protein structural fluctuations within a given substate structure cause the transition frequency to vary (spectral diffusion) (26–31). The landscape minimum associated with each substate is relatively shallow and has many local minima separated by small barriers (see Fig. 4). Transitions among these minima produce structural fluctuations on various time scales. At sufficiently long time, all structures associated with a substate are sampled and spectral diffusion is complete. All frequencies associated with the absorption band have been accessed. The 2D line shape goes from elongated to symmetrical. Fig. 2A shows that, by 48 ps, spectral diffusion is essentially complete. Here, we focus our analysis exclusively on the conformational switching; the protein structural dynamics that give rise to spectral diffusion of the A substates of MbCO have been reported previously (14, 29, 31).
Fig. 4.
Schematic illustration of a portion of the energy landscape of a protein, showing two substates and the finer energy landscape that exist in each of the minima. Transitions among the minima in one substate are responsible for structural fluctuations about the substate minimum. Transitions from one substate to another represent distinct structural configurational changes of the protein.
To quantitatively extract the time constant for the conformational switching from the 2D-IR spectra, the integrated peak volumes are fit to obtain the population of each species. The kinetic model described previously is used (15, 17, 20). It is possible to calculate the time evolution of the 2D spectrum including spectral diffusion and chemical exchange by using response function theory (20, 22, 23). However, in a detailed theoretical and experimental study, it was demonstrated that the exchange rate can be extracted by using the simpler method that is used here (20). Because spectral diffusion does not change the peak volumes, only their shapes, at each Tw the peak volumes were determined by fitting all of the peaks to 2D Gaussian functions (17, 20). The resulting fits provide the volume of each peak at each Tw. The conformational exchange causes the original peaks to decrease in volume and the cross peaks to increase in volume. In addition, the vibrational lifetime of the CO stretch, T1, causes all of the peaks to decrease in volume. In previous applications of the 2D-IR vibrational echo chemical-exchange method to small molecules in liquids, it was also necessary to account for the orientational relaxation of the species (15, 17, 20). Here, the protein is so large that the slow orientational relaxation can be neglected.
Eq. 1 shows the kinetic scheme.
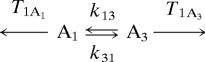 |
By using IR pump–probe experiments, the vibrational lifetimes, T1A1 and T1A3, were measured. The lifetimes are 25 and 19 ps, respectively. Because the system is in thermal equilibrium, the rate of A1 going to A3, k13[A1], has to equal the rate of A3 going to A1, k31[A3]. The ratio of substates determined from FT-IR and IR pump–probe experiment is [A1]/[A3] = 0.9. The substate switching time constant from A1 to A3 is τ13 = 1/k13. Therefore, in fitting the data, there is only one adjustable parameter, the substate switching time, τ13. There are eight peaks, four in the 0–1 region and four in the 1–2 region. The details of the fitting procedure have been presented previously (17, 20). The time-dependent volumes of all eight peaks are fit by using the single adjustable parameter, τ13.
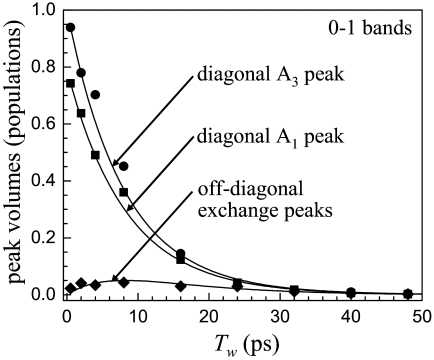
Fig. 2B shows calculated 2D spectra by using the known input parameters and the results of fitting τ13. Both the measured and the generated spectra are normalized by making the largest peak equal to unity at each time. The calculated 2D spectra are in good agreement with the time-dependent 2D spectra for 0–1 and 1–2 transition regions. The experimental diagonal and off-diagonal peak volumes for the 0–1 region of the spectra are plotted in Fig. 3. The solid lines through the data points are obtained from the fitting procedure with τ13 as the single adjustable parameter. The data from the 1–2 transition region can be reproduced with the identical parameters used for the 0–1 region. The fact that both the 0–1 and 1–2 regions can be fit with the same value of τ13 demonstrates that the thermal equilibrium of the system is not perturbed by vibrational excitation of the CO stretch (17). The results of the fitting yield τ13 = 47 ± 8 ps. Within experimental error, τ13 = τ31.
Fig. 3.
Peak volume data obtained from the fitting of the 2D-IR spectra of L29I-CO for the 0–1 transition region. The diagonal A1 and A3 bands as well as the off-diagonal peaks that grow in because of substate switching are fit with a single adjustable, the substate switching time, τ13 = 1/k13. The solid curves are the results of the fits, which yield τ13 = 47 ps.
Although Mb is one of the most intensively studied proteins and a great deal of attention has been focused on the nature and dynamics of the substates (1–3, 7–9), previously it has not been possible to measure the conformational switching between the A1 and A3 substates. The growth of the cross peaks in the 2D-IR vibrational echo spectra provides a direct observation of the conformational exchange. The interconversion time of 47 ps is faster than, but consistent with, the rough estimates that were made previously (13, 14). Because the 3D structure of L29I mutant Mb has not been reported, the impact of the amino acid substitution on the protein structure and dynamics is not clear. However, the peak frequencies of the CO absorption bands of L29I are virtually identical to the peak frequencies of wild-type Mb. The frequencies of the CO peaks in the IR spectrum are very sensitive to the protein structure. The fact that L29I-CO and MbCO have the same peak frequencies demonstrates that the perturbation of the structure of the heme pocket is small.
By using a combination of vibrational echo experiments and MD simulations, aspects of the structural change associated with the conformational switching between the A1 and A3 substates were established (14). The structural difference between the substates that is most directly involved in the difference in frequency of the A1 and A3 absorption peaks involves the imidazole side group of the distal histidine (Nε protonated) rotating about the Cβ Cγ (methylene carbon-imidazole ring carbon) bond. Movement of the distal histidine, which brings the Nε
Cγ (methylene carbon-imidazole ring carbon) bond. Movement of the distal histidine, which brings the Nε H into closer proximity to the CO, gives rise to the A3 substate (11, 14). The A1 substate has the distal histidine in a configuration that places the Nε
H into closer proximity to the CO, gives rise to the A3 substate (11, 14). The A1 substate has the distal histidine in a configuration that places the Nε H further from the CO (14). In the absence of interactions between the imidazole and any other moiety, it would be possible for the imidazole side group of a histidine amino acid to rotate about the Cβ
H further from the CO (14). In the absence of interactions between the imidazole and any other moiety, it would be possible for the imidazole side group of a histidine amino acid to rotate about the Cβ Cγ bond without causing translation or reorientation of the remainder of the molecule. In the crowded environment of the heme pocket, this is not likely to be the case. Switching between the A1 and A3 substates changes the interaction of the His-64 imidazole side group with the heme-CO and other moieties (see Fig. 1A). The change in the interactions should cause the His-64 to slightly reorient and translate. The structures of the heme pocket for the A1 and A3 substates taken form the MD simulation are shown in figure 5 of ref. 14. In these figures, which are single snap shots of the A1 and A3 structures, it is evident that with the rotation of the imidazole ring by ≈40° around the Cβ
Cγ bond without causing translation or reorientation of the remainder of the molecule. In the crowded environment of the heme pocket, this is not likely to be the case. Switching between the A1 and A3 substates changes the interaction of the His-64 imidazole side group with the heme-CO and other moieties (see Fig. 1A). The change in the interactions should cause the His-64 to slightly reorient and translate. The structures of the heme pocket for the A1 and A3 substates taken form the MD simulation are shown in figure 5 of ref. 14. In these figures, which are single snap shots of the A1 and A3 structures, it is evident that with the rotation of the imidazole ring by ≈40° around the Cβ Cγ bond there is also a change in the alignment of the Cα
Cγ bond there is also a change in the alignment of the Cα Cβ bond relative to the heme. These considerations suggest that there is a change in the geometry of the E helix (see Fig. 1A) in going between the A1 and A3 substates.
Cβ bond relative to the heme. These considerations suggest that there is a change in the geometry of the E helix (see Fig. 1A) in going between the A1 and A3 substates.
That significant structural change occurs in the A1–A3 interconversion is supported by x-ray experiments. The high-resolution crystal structure of MbCO that contains two conformations has enabled modeling of the structure of A1 and A3 substates (11, 32). Although the distal histidine has a critical role in determining the substates of Mb, structural comparison between the A1 and A3 substates shows that the A3 substate contains an additional cavity, Xe3, and another transient cavity found in simulations (32). Xe is used as a probe to identify the locations of cavities in proteins (33, 34). In Mb crystals, four Xe atoms (Xe1, Xe2, Xe3, and Xe4) occupy cavities, which may be involved in gas ligand migration (32, 33). The Xe3 site, which is near the surface and far from the iron atom, involves Trp-7 and is located between helices E and H (33). The existence of Xe3 in the A3 substate but not in the A1 substate demonstrates that the difference in the substates is significantly more than the rotation of the imidazole side group of the distal histidine.
The fast time constant for interconversion, 47 ps, is in line with the structural difference between the A1 and A3 substates. Incoherent quasielastic neutron scattering experiments on native bovine α-lactalbumin observed collective motions on the tens of picosecond to 100-ps time scale (35). The correlation length for such fluctuations was reported to be 18 Å (35). The switching time observed here falls into this time range, which is consistent with a reconfiguration of the E helix in the interconversion between the A1 and A3 substates.
Concluding Remarks
NMR techniques probe protein motions in the microsecond, millisecond, and longer time scales (36). Conformational switching processes studied by NMR involve large structural changes that occur, for example, in enzyme catalytic processes (37). Such structural changes require the reconfiguration of many amino acids, helices, and various protein structures. To place in a broader context the time-dependent measurements of substate switching presented here, a single elementary structural change, it is useful to discuss the kinetics of large-scale structural changes in terms of the ideas of linear response. Linear response theory describes how thermal fluctuations and elementary steps bring a system into a new configuration.
The concept of linear response comes from the fluctuation dissipation theorem (38). It states that a system in thermodynamic equilibrium has a response to a small perturbation with a time dependence determined by the equilibrium fluctuations of the system. The time-dependent fluorescence Stokes shift experiment is a well studied example of relaxation to a new equilibrium structure after a perturbation (39). In the experiment, a solute molecule with a small or zero permanent dipole moment in the ground electronic state is excited to the first excited state that has a large dipole moment. The local solvent structure will evolve to produce a net alignment of the solvent dipoles relative to the newly created large solute dipole. The orientational alignment of the solvent dipoles occurs in the same manner as thermal equilibrium orientational relaxation. (Translational relaxation will also occur.) Before the generation of the solute dipole, the solvent molecules are executing angular random walks (orientational diffusion). The sudden presence of the solute dipole slightly biases the random walks toward alignment. The alignment of a single solvent dipole can be viewed as an elementary step in the restructuring of the solvent. The solvation of the solute dipole requires the combined response of many solvent molecules.
Fig. 4 displays a schematic of a portion of the energy landscape of a protein having two substates. Each substate minimum is actually a broad rough landscape with many local minima separated by barriers of varying heights. Transitions between these minimum are responsible for protein structural fluctuations about a specific structure associated with a particular substate minimum (1–3). These fluctuations can vary in time scales from subpicosecond to tens and hundreds of picoseconds to much longer (1–3). The fluctuations range from the fastest motions involving just a few atoms to low-frequency acoustic-type modes of the entire protein. The distinction between structural fluctuations that come from making transitions among the minima on the landscape of a particular substate and transitions between substates is in some sense operational. A substate has a structural aspect that is qualitatively distinct. Fluctuations make interconversions among substates possible by sampling the energy landscape in a particular substate and bringing the protein to the transition state between substates.
When a protein experiences a perturbation such as substrate binding or a temperature jump that induces folding or unfolding, in accord with linear response, it will respond to the perturbation by fluctuation-driven elementary steps (substate changes) that can occur in the absence of the perturbation. However, the sampling of the substates will be biased by the perturbation, and the protein will relax to a new structure. The protein is always executing a multidimensional walk among substates. The walk will be skewed by the perturbation. The skewing can result from the shifting of substate minima and barriers. The relaxation to the new structure can be slow because it requires many elementary steps. A slow response to a perturbation results from structural fluctuation sampling that produces elementary steps, which in turn combine to produce major restructuring.
Here, by using ultrafast 2D-IR vibrational echo chemical-exchange spectroscopy, we have measured the time dependence of a single elementary step, the A1–A3 substate switching. The time constant for the substate switching is 47 ps. The concept of conformational substates in proteins was introduced in 1974 (3). In 1987, MD simulations of Mb for 300 ps confirmed the existence of multiple conformational states (40). The rapid advance of computer technology makes simulations of protein dynamics for much longer time scales readily doable. The A1–A3 substate switching time reported here will be an important target for future MD simulations because the barrier crossing will need to be simulated accurately. Simulations will determine whether the dynamics of real systems undergoing an elementary structural change can be reproduced, and they will provide details of the structural changes that accompany the A1–A3 interconversion.
Materials and Methods
Expression and purification of the mutant sperm whale Mb L29I were performed as described in ref. 41. The CO forms of mutant Mb was prepared according to published protocols (14). For both the linear FT-IR and vibrational echo measurements, ≈20 μl of the sample solution was placed in a sample cell with CaF2 windows and a 50-μm Teflon spacer. The 2D-IR vibrational echo experiments were described briefly above, and full details have been published previously (17, 19, 29, 42).
Acknowledgments.
We thank Prof. John S. Olson (Rice University, Houston, TX) for providing the L29I Mb mutant protein. This work was supported by National Institutes of Health Grant 2 R01 GM-061137-05 and Air Force Office of Scientific Research Grant F49620-01-1-0018. H.I. was supported by a fellowship from the Human Frontier Science Program.
Footnotes
The authors declare no conflict of interest.
References
- 1.Frauenfelder H, Parak F, Young RD. Conformational substates in proteins. Ann Rev Biophys Biophys Chem. 1988;17:451–479. doi: 10.1146/annurev.bb.17.060188.002315. [DOI] [PubMed] [Google Scholar]
- 2.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 3.Austin RH, et al. Activation-energy spectrum of a biomolecule: Photodissociation of carbonmonoxy myoglobin at low-temperatures. Phys Rev Lett. 1974;32:403–405. [Google Scholar]
- 4.Case DA, Karplus M. Dynamics of ligand-binding to heme-proteins. J Mol Biol. 1979;132:343–368. doi: 10.1016/0022-2836(79)90265-1. [DOI] [PubMed] [Google Scholar]
- 5.Schnell JR, Dyson HJ, Wright PE. Structure, dynamics, and catalytic function of dihydrofolate reductase. Annu Rev Biophys Biomol Struct. 2004;33:119–140. doi: 10.1146/annurev.biophys.33.110502.133613. [DOI] [PubMed] [Google Scholar]
- 6.Oliveberg M, Wolynes PG. The experimental survey of protein-folding energy landscapes. Q Rev Biophys. 2005;38:245–288. doi: 10.1017/S0033583506004185. [DOI] [PubMed] [Google Scholar]
- 7.Ansari A, et al. Rebinding and relaxation in the myoglobin pocket. Biophys Chem. 1987;26:337–355. doi: 10.1016/0301-4622(87)80034-0. [DOI] [PubMed] [Google Scholar]
- 8.Tian WD, Sage JT, Champion PM. Investigation of ligand association and dissociation rates in the “open” and “closed” states of myoglobin. J Mol Bio. 1993;233:155–166. doi: 10.1006/jmbi.1993.1491. [DOI] [PubMed] [Google Scholar]
- 9.Muller JD, McMahon BH, Chen EYT, Sligar SG, Nienhaus GU. Connection between the taxonomic substates of protonation of histidines 64 and 97 in carbonmonoxy myoglobin. Biophys J. 1999;77:1036–1051. doi: 10.1016/s0006-3495(99)76954-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li TS, Quillin ML, Phillips GN, Jr, Olson JS. Structural determinants of the stretching frequency of CO bound to myoglobin. Biochemistry. 1994;33:1433–1446. doi: 10.1021/bi00172a021. [DOI] [PubMed] [Google Scholar]
- 11.Vojtechovsky J, Chu K, Berendzen J, Sweet RM, Schlichting I. Crystal structures of myoglobin-ligand complexes at near atomic resolution. Biophys J. 1999;77:2153–2174. doi: 10.1016/S0006-3495(99)77056-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang F, Phillips GN., Jr Crystal structures of CO-, deoxy- and met-myoglobins at various pH values. J Mol Biol. 1996;256:762–774. doi: 10.1006/jmbi.1996.0123. [DOI] [PubMed] [Google Scholar]
- 13.Johnson JB, et al. Ligand binding to heme proteins. 6. Interconversion of taxonomic substates in carbonmonoxymyoglobin. Biophys J. 1996;71:1563–1573. doi: 10.1016/S0006-3495(96)79359-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merchant KA, et al. Myoglobin-CO substate structures and dynamics: Multidimensional vibrational echoes and molecular dynamics simulations. J Am Chem Soc. 2003;125:13804–13818. doi: 10.1021/ja035654x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng J, Kwak K, Xie J, Fayer MD. Ultrafast carbon-carbon single bond rotational isomerization in room-temperature solution. Science. 2006;313:1951–1955. doi: 10.1126/science.1132178. [DOI] [PubMed] [Google Scholar]
- 16.Kim YS, Hochstrasser RM. Chemical exchange 2D IR of hydrogen-bond making and breaking. Proc Natl Acad Sci USA. 2005;102:11185–11190. doi: 10.1073/pnas.0504865102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng J, et al. Ultrafast dynamics of solute-solvent complexation observed at thermal equilibrium in real time. Science. 2005;309:1338–1343. doi: 10.1126/science.1116213. [DOI] [PubMed] [Google Scholar]
- 18.Morikis D, Champion PM, Springer BA, Sligar SG. Resonance Raman investigations of site-directed mutants of myoglobin: Effects of distal histidine replacement. Biochemistry. 1989;28:4791–4800. doi: 10.1021/bi00437a041. [DOI] [PubMed] [Google Scholar]
- 19.Park S, Kwak K, Fayer MD. Ultrafast 2D-IR vibrational echo spectroscopy: A probe of molecular dynamics. Laser Phys Lett. 2007;4:704–718. [Google Scholar]
- 20.Kwak K, Zheng J, Cang H, Fayer MD. Ultrafast two-dimensional infrared vibrational echo chemical exchange experiments and theory. J Phys Chem B. 2006;110:19998–20013. doi: 10.1021/jp0624808. [DOI] [PubMed] [Google Scholar]
- 21.Zheng J, Kwak K, Chen X, Asbury JB, Fayer MD. Formation and dissociation of intra-intermolecular hydrogen-bonded solute-solvent complexes: Chemical exchange two-dimensional infrared vibrational echo spectroscopy. J Am Chem Soc. 2006;128:2977–2987. doi: 10.1021/ja0570584. [DOI] [PubMed] [Google Scholar]
- 22.Sanda F, Mukamel S. Stochastic simulation of chemical exchange in two dimensional infrared spectroscopy. J Chem Phys. 2006;125 doi: 10.1063/1.2205367. 014507. [DOI] [PubMed] [Google Scholar]
- 23.Sanda F, Mukamel S. Anomalous lineshapes and aging effects in two-dimensional correlation spectroscopy. J Chem Phys. 2007;127:154107. doi: 10.1063/1.2793786. [DOI] [PubMed] [Google Scholar]
- 24.Rector KD, et al. Vibrational anharmonicity and multilevel vibrational dephasing from vibrational echo beats. J Chem Phys. 1997;106:10027. [Google Scholar]
- 25.Golonzka O, Khalil M, Demirdoven N, Tokmakoff A. Vibrational anharmonicities revealed by coherent two-dimensional infrared spectroscopy. Phys Rev Lett. 2001;86:2154–2157. doi: 10.1103/PhysRevLett.86.2154. [DOI] [PubMed] [Google Scholar]
- 26.Mukamel S. Multidimensional femtosecond correlation spectroscopies of electronic and vibrational excitations. Ann Rev Phys Chem. 2000;51:691–729. doi: 10.1146/annurev.physchem.51.1.691. [DOI] [PubMed] [Google Scholar]
- 27.Mukamel S. Principles of Nonlinear Optical Spectroscopy. New York: Oxford Univ Press; 1995. [Google Scholar]
- 28.Finkelstein IJ, Ishikawa H, Kim S, Massari AM, Fayer MD. Substrate binding and protein conformational dynamics measured via 2D-IR vibrational echo spectroscopy. Proc Natl Acad Sci USA. 2007;104:2637–2642. doi: 10.1073/pnas.0610027104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finkelstein IJ, et al. Probing dynamics of complex molecular systems with ultrafast 2D IR vibrational echo spectroscopy. Phys Chem Chem Phys. 2007;9:1533–1549. doi: 10.1039/b618158a. [DOI] [PubMed] [Google Scholar]
- 30.Ishikawa H, Kim S, Kwak K, Wakasugi K, Fayer MD. Disulfide bond influence on protein structural dynamics probed with 2D-IR vibrational echo spectroscopy. Proc Natl Acad Sci USA. 2007;104:19309–19314. doi: 10.1073/pnas.0709760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishikawa H, et al. Neuroglobin dynamics observed with ultrafast 2D-IR vibrational echo spectroscopy. Proc Natl Acad Sci USA. 2007;104:16116–16121. doi: 10.1073/pnas.0707718104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teeter MM. Myoglobin cavities provide interior ligand pathway. Protein Sci. 2004;13:313–318. doi: 10.1110/ps.03334304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tilton RF, Jr, Kuntz ID, Jr, Petsko GA. Cavities in proteins: Structure of a metmyoglobin xenon complex solved to 1.9 Å. Biochemistry. 1984;23:2849–2857. doi: 10.1021/bi00308a002. [DOI] [PubMed] [Google Scholar]
- 34.Doukov TI, Blasiak LC, Seravalli J, Ragsdale SW, Drennan CL. Xenon in and at the end of the tunnel of bifunctional carbon monoxide dehydrogenase/acetyl-coa synthase. Biochemistry. 2008;47:3474–3483. doi: 10.1021/bi702386t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bu Z, et al. A view of dynamics changes in the molten globule native folding step by quasielastic neutron scattering. J Mol Bio. 2000;301:525–536. doi: 10.1006/jmbi.2000.3978. [DOI] [PubMed] [Google Scholar]
- 36.Palmer AG., III Probing molecular motion by NMR. Curr Opin Struct Biol. 1997;7:732–737. doi: 10.1016/s0959-440x(97)80085-1. [DOI] [PubMed] [Google Scholar]
- 37.Boehr DD, Dyson HJ, Wright PE. An NMR perspective on enzyme dynamics. Chem Rev. 2006;106:3055–3079. doi: 10.1021/cr050312q. [DOI] [PubMed] [Google Scholar]
- 38.Kubo R, Toda M, Hashitusme N. Statistical Physics II: Nonequilibrium Statistical Mechanics. New York: Springer; 1985. [Google Scholar]
- 39.Fleming GR, Mihnaeng C. Chromophore-solvent dynamics. Annu Rev Phys Chem. 1996;47:109–134. [Google Scholar]
- 40.Elber R, Karplus M. Multiple conformational states of proteins: A molecular dynamics analysis of myoglobin. Science. 1987;235:318–321. doi: 10.1126/science.3798113. [DOI] [PubMed] [Google Scholar]
- 41.Springer BA, Sligar SG. High-level expression of sperm whale myoglobin in Escherichia coli. Proc Natl Acad Sci USA. 1987;84:8961–8965. doi: 10.1073/pnas.84.24.8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng J, Kwak K, Fayer MD. Ultrafast 2D IR vibrational echo spectroscopy. Acc Chem Res. 2007;40:75–83. doi: 10.1021/ar068010d. [DOI] [PubMed] [Google Scholar]