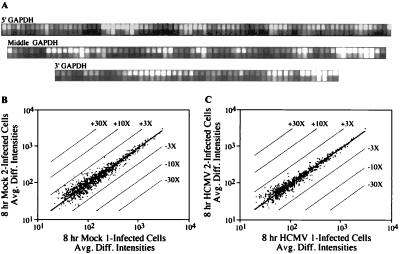
Figure 1.
Characterization of RNA target samples and reproducibility of array-based hybridization results. (A) Probe pairs (82, 68, and 51 pairs) were used to interrogate the 5′, middle, and 3′ portions of the glyceraldehyde-3-phosphate dehydrogenase mRNA, which is expressed constitutively in fibroblasts. (B and C) Plots comparing the average difference intensities in fluorescent signal (Avg. Diff. Intensities) of the 20 probe pairs interrogating each of the genes present in two independent experiments performed on the mock-infected cells (B) or cells at 8 h after infection (C). The parallel lines flanking the center diagonal line indicate 3-, 10-, and 30-fold changes in intensity. With the exception of the thombospondin 1 gene in the mock-infected control, all genes demonstrated an average difference in their fluorescent intensities of <3-fold.