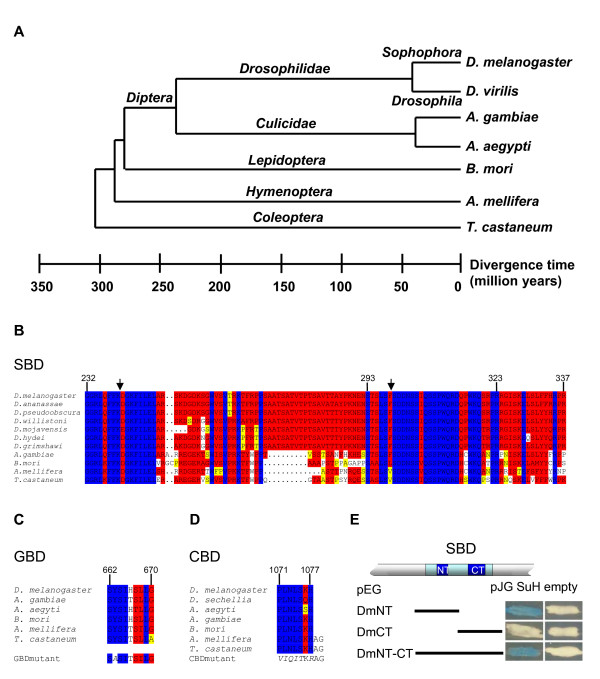
Figure 1.
Conservation of H structural domains in various insects. A) Evolutionary relationship between insect orders and families, including Drosophilids, Nematocera Diptera, Lepidoptera, Hymenoptera and Coleoptera. Estimated distance is given in million years. B) Alignments of the SBD, (C) the GBD, (D) the CBD of H orthologues from different insects. 13 different Drosophilids were analyzed (see also Additional file 1); only those sequences are depicted that are different from D. melanogaster or other closely related species. Both, A. aegypti and A. gambiae were analyzed; A. aegypti is only shown if different from A. gambiae. Numbers above the domains correspond to D.m.H amino acid sequence. Below the GBD and CBD, the experimentally introduced mutations are shown. Identical residues are marked in blue; red shows highly conserved and yellow similar residues; dots mark gaps. E) Qualitative analysis of D. melanogaster SBD binding to Su(H). Yeast two-hybrid assay to demonstrate interaction between Su(H) [pJG-Su(H)] and SBD or parts thereof. pJG empty vector served as control. Dm-NT-CT overlaps the SBD, Dm-NT includes the N-terminal and Dm-CT the C-terminal portion. Positive interactions are recognized by the blue color caused by transcriptional activation of a lacZ-reporter gene.