TABLE 2.
Autocorrelation analysis (see Appendix) of the binary mixing samples
| R | Y† | Bias† (Y-R) |
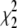 † †
|
〈τr〉* (μs) | 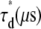 |
β* |
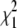 * *
|
|---|---|---|---|---|---|---|---|
| 0 | 0.000 ± 0.020 | 0.000 | 0.46 | 7.3 ± 0.3 | 217.2 ± 5.6 | 0.585 ± 0.009 | 0.47 |
| 0.125 | 0.146 ± 0.012 | 0.021 | 0.47 | 6.6 ± 0.3 | 223.3 ± 9.1 | 0.571 ± 0.016 | 0.44 |
| 0.25 | 0.247 ± 0.014 | −0.003 | 0.49 | 6.3 ± 0.2 | 231.5 ± 7.5 | 0.549 ± 0.012 | 0.47 |
| 0.375 | 0.354 ± 0.011 | −0.021 | 0.53 | 5.9 ± 0.3 | 237.8 ± 9.8 | 0.527 ± 0.021 | 0.53 |
| 0.5 | 0.480 ± 0.008 | −0.020 | 0.85 | 5.2 ± 0.2 | 246.4 ± 9.8 | 0.516 ± 0.013 | 0.74 |
| 0.625 | 0.639 ± 0.006 | 0.014 | 0.88 | 4.2 ± 0.0 | 252.0 ± 2.6 | 0.493 ± 0.012 | 0.85 |
| 0.75 | 0.726 ± 0.014 | −0.024 | 1.31 | 3.6 ± 0.1 | 253.7 ± 5.8 | 0.494 ± 0.014 | 1.14 |
| 0.875 | 0.874 ± 0.006 | −0.001 | 1.41 | 2.8 ± 0.0 | 263.4 ± 2.2 | 0.511 ± 0.011 | 1.41 |
| 1 | 0.999 ± 0.008 | −0.001 | 1.71 | 2.1 ± 0.1 | 265.3 ± 10.0 | 0.548 ± 0.034 | 1.36 |
The values 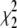 and
and 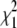 measure the goodness of the fit for the two-component and one-component models, respectively (42).
measure the goodness of the fit for the two-component and one-component models, respectively (42).
From the one-component model.
From the two-component model.
R, mixing molar fraction of dsDNA to total DNA; Y, fraction of Cy5-labeled oligo forming duplex; Bias, Y-R; 〈τr〉, mean relaxation time; τd, diffusion time; β, stretch parameter.
