TABLE 1.
Data collection and refinement statistics
| Statistic | Type 1 | Type 2 | ||
|---|---|---|---|---|
| Data collection | ||||
| Space group | P1 | P1 | ||
| Unit cell dimensions (Å, degree) | a = 42.36, b = 65.84, c = 71.14 | a = 43.20, b = 63.72, c = 73.26 | ||
| α = 66.40, β = 89.93, γ = 90.04 | α = 66.64, β = 89.99, γ = 90.30 | |||
| Spacing (Å) | 20.0 − 1.80 | 1.86 − 1.80 | 29.25 − 2.00 | 2.07 − 2.00 |
| No. of measured reflections | 244,351 | 182,859 | ||
| No. of unique reflections | 62,779 | 47,224 | ||
| Average redundancy | 3.9 | 3.7 | 3.9 | 3.7 |
| Completeness (%) | 96.3 | 95.1 | 97.4 | 93.6 |
| Rmerge (I)* | 0.064 | 0.201 | 0.038 | 0.110 |
| Average I/σ (I) | 17.1 | 5.5 | 21.0 | 4.8 |
| Solvent content (%)/VM | 50.1/2.47 | 51.0/2.47 | ||
| Refinement | ||||
| No. of reflections (%) | 62,708 (96.0) | 47,200 (97.2) | ||
| No. of reflections in working set (%) | 59,547 (91.3) | 44,801 (92.4) | ||
| No. of reflections in test set (%) | 3161 (4.8) | 2399 (4.9) | ||
| R† | 0.237 | 0.249 | ||
| Free R‡ | 0.316 | 0.304 | ||
| No. of peptide chains | 4 | 4 | ||
| No. of protein/water atoms | 5118/1460 | 5096/361 | ||
| RMSD bond length (Å) | 0.008 | 0.008 | ||
| RMSD bond angle (degree) | 1.1 | 1 | ||
| Average B factor | 42.4 | 52.9 | ||
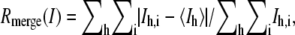 where Ih,i is the intensity of the ith observation of the reflection h.
where Ih,i is the intensity of the ith observation of the reflection h.
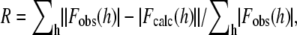 where Fobs(h) and Fobs(h) are the observed and calculated structure factors, respectively.
where Fobs(h) and Fobs(h) are the observed and calculated structure factors, respectively.
Free R is calculated for randomly selected 5% of the reflection data, which were not included in the refinement.
