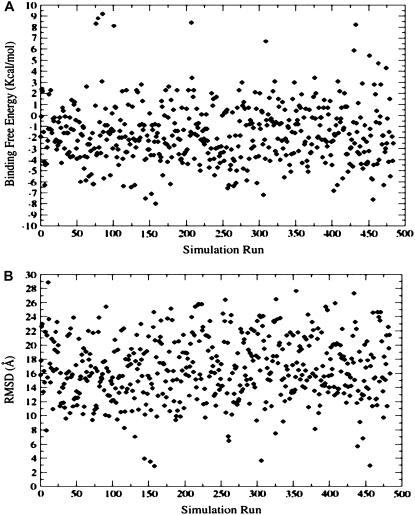
FIGURE 2.
(A) The MM/GBSA binding free energies as a function of simulation run obtained from 500 flexible docking simulations. (B) The RMSD values of the docked conformations are reported in the reference to the lowest energy structure of the p-S8 inhibitor corresponding to the folding mode of inhibition and reflect two structural clusters of low-energy docking solutions.