Fig. 4.
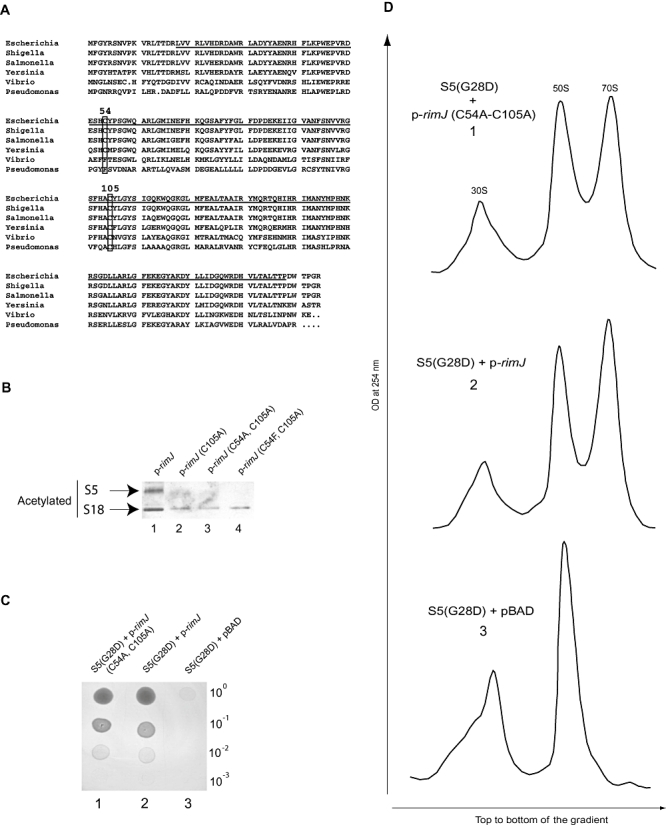
RimJ can suppress defects associated with S5(G28D) independent of its acetyltransferase activity. Overexpression of wild-type RimJ in the ΔrimJ or S5(G28D) background by induction of p-rimJ. Mutant RimJ proteins were overexpressed from p-rimJ(C105A), p-rimJ(C54A, C105A) and p-rimJ(C54F, C105A). Numbers and letters after the cysteines (C) correspond to positions in E. coli RimJ sequence that have been mutated to alanine (A) or phenylalanine (F). A. Multiple alignment of RimJ sequences and positions of C54 and C105. Sequence comparisons of RimJ from various prokaryotic species. The NAT domain in E. coli RimJ sequence is underlined. C54 and C105 residues are boxed. The RimJ sequences used in sequence alignment and their accession numbers in parenthesis are as follows. Escherichia: Escherichia coli (NP_415584), Shigella: Shigella flexneri (P0A950), Salmonella: Salmonella enterica (NP_455660), Yersinia: Yersinia pestis (CAL20676), Vibrio: Vibrio cholerae (YP_001216873), Pseudomonas: Pseudomonas aeruginosa (YP_791309). B. Western analysis for acetylated proteins in 30S subunit proteins (TP30) obtained from the ΔrimJ strain (CWZ387) with overexpression of various constructs: (1) p-rimJ (2) p-rimJ(C105A) (3) p-rimJ(C54A, C105A) (4) p-rimJ(C54F, C105A). C. 10-fold serial dilutions of S5(G28D) with various constructs at the non-permissive temperature, 20°C: (1) p-rimJ(C54A, C105A) (2) p-rimJ (3) empty pBAD. D. Sucrose gradient sedimentation analysis of ribosomes from S5(G28D) grown at the non-permissive temperature, 20°C, with the constructs: (1) p-rimJ(C54A, C105A) (2) p-rimJ (3) empty pBAD. Positions of the 30S and 50S subunits and 70S ribosomes are indicated.
