Fig. 5.
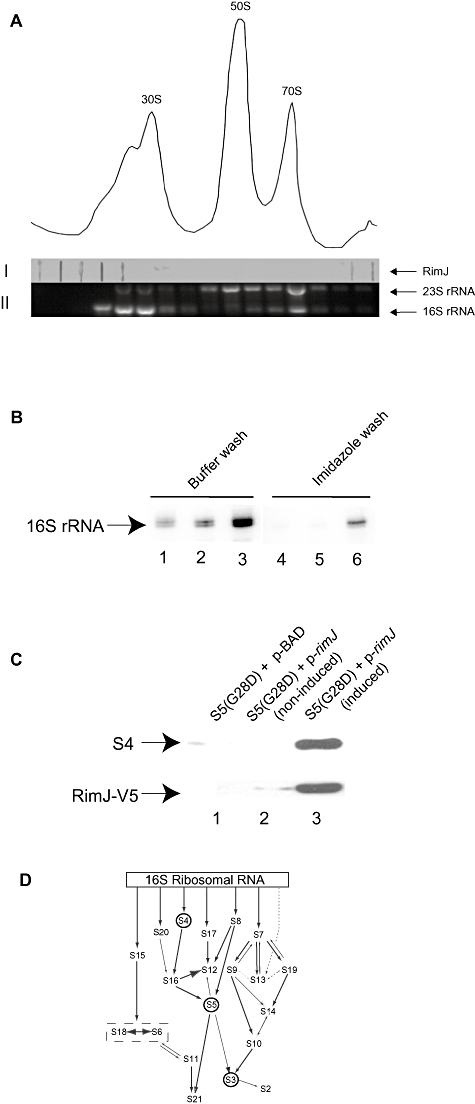
RimJ associates with assembling 30S subunits in vivo. A. Analysis of ribosomal profile of S5(G28D) grown at the non-permissive temperature with induction of p-rimJ. Fractions from across this gradient were analysed by (I) slot-blot for RimJ or (II) gel electrophoresis for 23S rRNA and 16S rRNA to determine location of ribosomal particles. B. Primer extension analysis of 16S rRNAs extracted from immunoprecipitated RNPs with pBAD (Lanes 1, 4), non-induced p-rimJ (Lanes 2, 5) and induced p-rimJ (Lanes 3, 6) in S5(G28D) cells. Ni-NTA beads were washed with B-150 buffer (Lanes 1–3) or imidazole (Lanes 4–6). C. Western analysis of RNPs immunoprecipitated with pBAD (Lane 1), non-induced p-rimJ (Lane 2) and induced p-rimJ (Lane 3) in S5(G28D) cells for S4 and RimJ-V5 (indicated by arrows). D. Modified in vitro assembly map for 30S subunits (Grondek and Culver, 2004). Arrows indicate dependency between components for association with 16S rRNA. Positions of S4, S5 and S3 are indicated by circles.
