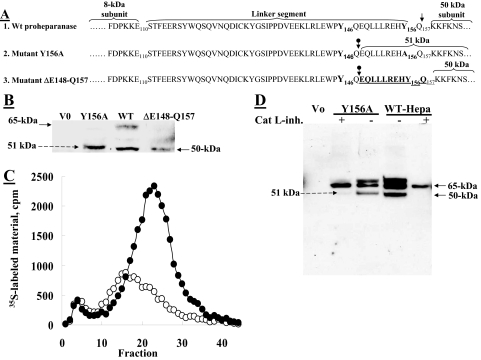
FIGURE 5.
A 10-amino acid peptide at the C terminus of the linker segment is critical for proheparanase activation. A, schematic presentation of WT proheparanase undergoing processing at Y156Q (↓), yielding the accurate 50-kDa subunit (1). Point-mutated (Y156A) proheparanase, yielding a 51-kDa polypeptide composed of the 50-kDa subunit conjugated to a 10-amino acid peptide at the C terminus of the linker (2), due to cleavage inside the linker segment at the cathepsin L motif Y146Q (circle with down arrow). Deletion mutant, ΔGlu148–Gln157, spanning the 10-amino acid peptide at the C terminus of the linker segment, including the Y156Q cleavage site, which undergoes processing at Y146Q, generating the proper 50-kDa subunit (3). B, JAR cells were stably transfected with empty plasmid (V0), pcDNA encoding the full-length heparanase cDNA (WT), pcDNA encoding the point mutated heparanase cDNA (Y156A), or pcDNA encoding the deletion mutant (ΔGlu148–Gln157). Cell lysates were subjected to Western blot analysis using anti-heparanase pAb 1453. C, JAR cells transfected with the Y156A (○) or ΔGlu148–Gln157 (•) heparanase mutants were assayed for heparanase enzymatic activity (7 h, pH 6.0, 37 °C) on sulfate labeled ECM. D, JAR cells were either mock transfected (Vo) or transiently transfected with wild-type heparanase (WT) or the Y156A point-mutated heparanase. The cells were grown (24 h) in the absence or presence of 0.72 μm cathepsin L inhibitor I (catalog number 219421, Calbiochem), extracted (1% Nonidet P-40, 10 mm EDTA in PBS supplemented with protease inhibitors), and subjected to Western blot analysis of heparanase, applying pAb 1435.