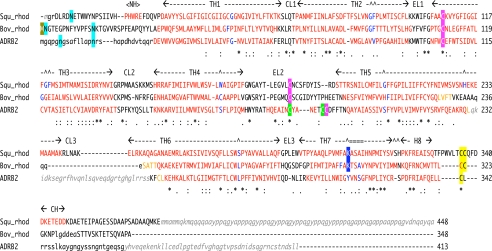
FIGURE 1.
Structural sequence alignment of squid rhodopsin, bovine rhodopsin and β2AR. The structural alignment was based on the 3D-Coffee alignment (4). Residues on helix regions are colored red, and residue(s) of helix bending are colored in blue. Transmembrane helical regions (TH1–TH7) and helix H8 with extracellular (EL1–EL3) and cytoplasmic (CL1–CL3) loops, and each helix in N- and C-terminal tails (NH and CH) are indicated. Posttranslational modifications are shaded by the following colors: cyan, N-glycosylation; a pair of pink or green, disulfide bridge(s); yellow, palmitoylated cysteine; blue, Schiff-based lysine with 11-cis-retinal; gold, N-terminal methionine acetylation. Residues indicated by small letters are not in models but in crystal protein samples, and residues indicated by small letters in italic gray do not exist in the crystal sample proteins due to expression processing, protease digestion, or protein engineering. Squ_rhod, squid rhodopsin (PDB code: 2ZIY in this study); Bov_rhod, bovine rhodopsin (1F88 (5) or 1GZM (17)); and ADRB2 (2RH1 (8)).