Fig. 3.
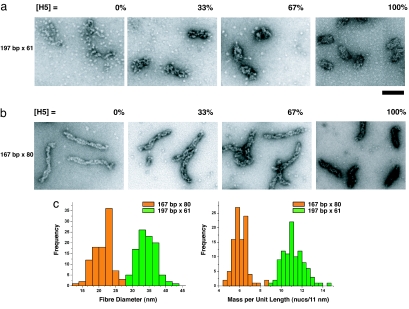
The 167- and 197-bp NRL arrays have different linker histone-dependent compaction pathways and different structures. (a and b) For EM analysis, the 167-bp × 80 and 197-bp × 61 nucleosome arrays were reconstituted with different concentrations of linker histone as in Fig. 2 and folded in 1.6 mM MgCl2. The percentage of linker histone H5 saturation (as derived from Fig. 2b) is indicated above each panel. The particles seen in the background of the EM micrographs are individual nucleosomes resulting from excess histone octamer bound to competitor DNA. (a) EM micrographs showing the folding pathway of the 197-bp NRL array from disordered puddles in [H5] = 0%, to the formation of regular 30-nm chromatin fibers at saturation of H5 binding ([H5] = 100%). (b) EM micrographs showing the folding pathway of the 167-bp NRL arrays from thin ladder-like structures formed by the stacking of nucleosome cores in [H5] = 0%, to the formation of thin, more twisted, fibers at saturation of H5 binding ([H5 = 100%]). (c) Histogram representations of the diameter and mass per unit length for the 197-bp × 61 (green) and 167-bp × 80 (orange) fully folded chromatin fibers saturated with H5. Ninety-nine measurements were taken from EM micrographs for each array. The average diameter and mass per unit length for the 167-bp NRL fibers is 21.3 nm (SD = 3.0) and 6.1 nucleosomes per 11 nm (SD = 0.74), and the average diameter and mass per unit length for the 197-bp NRL fibers is 34.3 nm (SD = 2.7) and 11.2 nucleosomes per 11 nm (SD = 1.0). (Scale bar: 100 nm.)