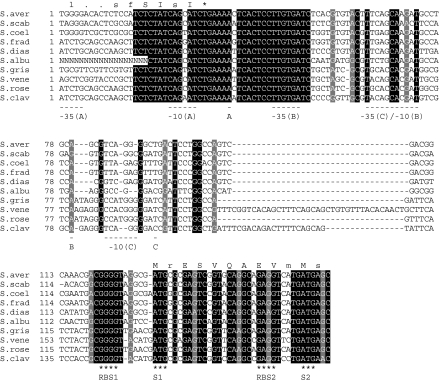
Fig. 2.
Alignment of ssgA promoter regions. Alignment was produced using the Boxshade program (www.ch.embnet.org/software/BOX_form.html). Only completely conserved nucleotides are shaded; nucleotides shaded in light grey refer to conserved purines. The two alternative start codons (S) for ssgA and their respective ribosome binding sites (RBS) are indicated below the aligned sequence. The two transcriptional start sites and their respective −35 and −10 recognition sequences from S. griseus (Yamazaki et al. 2003) and S. coelicolor (Traag et al. 2004) are underlined, where “A” refers to p1 from S. griseus, “B” to p1 from S. coelicolor or p2 from S. griseus, and “C” to p2 from S. coelicolor. Consensus amino acid sequences of the C-terminus of SsgR proteins and the N-terminus of SsgA proteins are given above the aligned DNA sequences. The TGA stop codon for ssgR is indicated with an asterisk