Abstract
Background
Branchiomotor neurons comprise an important class of cranial motor neurons that innervate the branchial-arch-derived muscles of the face, jaw and neck. They arise in the ventralmost progenitor domain of the rhombencephalon characterized by expression of the homeodomain transcription factors Nkx2.2 and Phox2b. Phox2b in particular plays a key role in the specification of branchiomotor neurons. In its absence, generic neuronal differentiation is defective in the progenitor domain and no branchiomotor neurons are produced. Conversely, ectopic expression of Phox2b in spinal regions of the neural tube promotes cell cycle exit and neuronal differentiation and, at the same time, induces genes and an axonal phenotype characteristic for branchiomotor neurons. How Phox2b exerts its pleiotropic functions, both as a proneural gene and a neuronal subtype determinant, has remained unknown.
Results
To gain further insights into the genetic program downstream of Phox2b, we searched for novel Phox2b-regulated genes by cDNA microarray analysis of facial branchiomotor neuron precursors from heterozygous and homozygous Phox2b mutant embryos. We selected for functional studies the genes encoding the axonal growth promoter Gap43, the Wnt antagonist Sfrp1 and the transcriptional regulator Sox13, which were not previously suspected to play roles downstream of Phox2b and whose expression was affected by Phox2b misexpression in the spinal cord. While Gap43 did not produce an obvious phenotype when overexpressed in the neural tube, Sfrp1 induced the interneuron marker Lhx1,5 and Sox13 inhibited neuronal differentiation. We then tested whether Sfrp1 and Sox13, which are down-regulated by Phox2b in the facial neuron precursors, would antagonize some aspects of Phox2b activity. Co-expression of Sfrp1 prevented Phox2b from repressing Lhx1,5 and alleviated the commissural axonal phenotype. When expressed together with Sox13, Phox2b was still able to promote cell cycle exit and neuronal differentiation, but the cells failed to relocate to the mantle layer and to extinguish the neural stem cell marker Sox2.
Conclusion
Our results suggest novel roles for Sfrp1 and Sox13 in neuronal subtype specification and generic neuronal differentiation, respectively, and indicate that down-regulation of Sfrp1 and Sox13 are essential aspects of the genetic program controlled by Phox2b in cranial motoneurons.
Background
Branchiomotor (bm) neurons comprise an important class of cranial motoneurons. They arise in the hindbrain and rostral cervical spinal cord from the ventralmost progenitor domain characterized by expression of the transcription factors Nkx2.2, Mash1 and Phox2b [1,2]. Targeted mutation in the mouse has shown that their development crucially depends on Phox2b whereas the inactivation of Nkx2.2 or Mash1 results in only mild defects of bm development [2-5]. In the absence of Phox2b, the differentiation of all bm neurons is arrested at an early stage and their precursors either die by apoptosis or switch fate and become serotonergic neurons. Conversely, Phox2b induces neurons with bm characteristics when misexpressed in the neural tube of chicken embryos [2,6]. Hence, Phox2b appears both necessary and sufficient for the implementation of a specific cranial motoneuronal phenotype. Loss and gain of function experiments also show that Phox2b has proneural activity [7] and promotes cell cycle exit and generic neuronal differentiation [8] (see for review [9]). The development of the facial bm (fbm) neurons that arise in rhombomere 4 (r4) has been particularly well studied in the Phox2b knockout embryos. A number of downstream target genes have been identified that depend on Phox2b for their expression in fbm neurons by examining expression of candidate genes and by a subtractive screening approach. They include generic markers for young post-mitotic neurons (Tubb3, Nfl, Ebf2, Math3) and genes more specifically associated with bm neuron differentiation (Phox2a, Islet1, Rgs4, Tbx20) ([4,10] and unpublished results).
Despite this wealth of information, only part of the genetic program that depends on Phox2b in fbm neurons has been identified. Indeed, the subtractive screening approach proved far from exhaustive since most previously identified downstream genes were not detected and none were found to be up-regulated in the mutants. Here we have used microarrays to compare the patterns of gene expression in fbm precursors from homozygous (Phox2bLacZ/LacZ) mutants, in which fbm differentiation is arrested, with that of heterozygous Phox2b (Phox2bLacZ/+) embryos, in which fbm neurons are phenotypically normal. In addition to genes already identified in previous work, a number of new candidates turned up in the screen, among which many were up-regulated in the mutants and might thus be repressed by Phox2b. Most of the down-regulated genes were panneuronally expressed in young post-mitotic neurons. They were thus expected to pop up in the screen, since fewer post-mitotic neurons are produced in the mutants, and were not analyzed further. We selected a few other genes for further study by gain of function experiments in chicken embryos: Gap43, whose expression, although panneuronal, was strikingly enhanced in post-mitotic fbm precursors; and two that were up-regulated in the mutants, Sfrp1 and Sox13. Overexpression of Sox13, whose function in neural development has not been studied so far, inhibited neuronal differentiation and the relocation to the mantle layer (ML) caused by Phox2b misexpression in the embryonic spinal cord, while that of Sfrp1 prevented the repression of the interneuronal marker Lhx1,5.
Results
Identification of genes regulated by Phox2b in facial branchiomotor precursors
Phox2b being an important regulator of the development of bm neurons, we wanted to further investigate its function in the specification of bm neurons by searching for novel genes regulated by Phox2b in fbm neuron precursors. We thus chose to compare gene expression in ventral r4 from heterozygous (Phox2bLacZ/+) and homozygous (Phox2bLacZ/LacZ) embryonic day 10.5 (E10.5) mutant embryos. At this stage, fbm differentiation is well underway in Phox2b+/+ and Phox2bLacZ/+ animals while no signs of bm differentiation are seen in the homozygous mutants [4]. Four pairs of Phox2bLacZ/+ and Phox2bLacZ/LacZ RNA samples were amplified from microdissected ventral r4, labeled with Cy3 or Cy5, mixed and hybridized to two mouse cDNA microarrays, a microarray of the 15 K NIA probe set representing approximately 12,000 different genes and a home-made NeuroDev microarray representing 2,000 genes chosen for their expression in developing nervous tissue. We adopted as criteria for putative Phox2b-regulated genes a significance analysis of microarray (SAM) score (see Materials and methods) below -3.5 or above 3.5 for genes up- or down-regulated in the mutants, respectively, and a false discovery rate of 5%. By these criteria, 51 genes were found to be down-regulated and 23 up-regulated in the homozygous mutants (Additional files 1 and 2). Surprisingly, only three genes showed a greater than twofold change in expression. However, out of the 12 genes we selected for validation by in situ hybridization (ISH), 10 were confirmed as being clearly differentially expressed, despite fold changes as low as 1.14 and 1.13 for two of them (see below). Part of the rather low signal ratios on microarrays may be due to the fact that the floor plate, which should not be affected in the mutants, was always included in the material used for RNA preparation.
Only two of the genes that scored as underexpressed in the mutants, Tubb3 and Nfl, had been known from previous work ([4] and unpublished data). We selected six for validation by ISH, which was done on sections through r4 from E10.5 Phox2bLacZ/+ and Phox2bLacZ/LacZ embryos. The expression of one of them, Sncg, was not noticeably affected in the mutants (not shown). The remaining five fell into two categories. App, coding for the amyloid beta (A4) precursor protein, Mapk8, also known as Jun N-terminal kinase (Jnk1), and Tubb3, the beta3-tubulin gene (already documented in previous work) are pan-neuronal genes expressed in all developing neurons. In the mutants, their expression was interrupted at the dorsal border of the normally Phox2b+ territory (Figure 1a–d). The small number of post-mitotic neurons that are generated in the homozygous mutants and locate to the ML, some of which are in the process of acquiring a serotonergic fate [1], thus do not appear to activate these genes that are otherwise pan-neuronal. Gap43, a major growth cone protein [11,12], and to a lesser extent Nfl, coding for the neurofilament light chain, differed from the others by the fact that, although pan-neuronal as well, they were more strongly expressed in the fbm precursors than in other young post-mitotic neurons. Their expression was extinguished in the mutant territory (Figure 1e–h).
Figure 1.
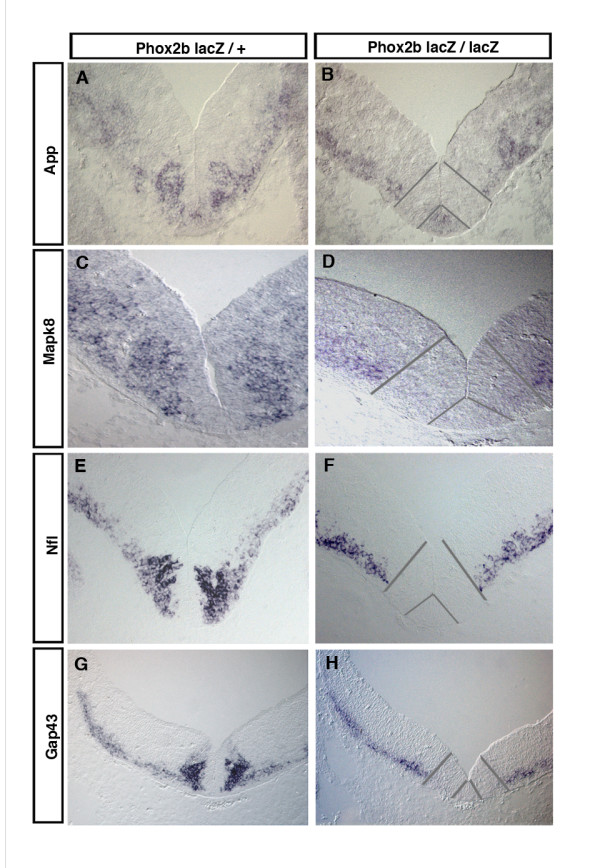
Expression patterns of a subset of genes down-regulated in ventral r4 of Phox2bLacZ/LacZ embryos. (a-h) In situ hybridizations on transverse sections of ventral r4 of Phox2bLacZ/+ (a, c, e, g) or Phox2bLacZ/LacZ (b, d, f, h) E10.5 mouse embryos. DIG-labeled riboprobes recognizing App (a, b), Mapk8 (c, d), Nfl (e, f) and Gap43 (g, h) mRNA were used. The normally Phox2b-expressing region as visualized by anti-beta-galactosidase staining on adjacent sections is delimited by gray bars.
In the absence of Phox2b, less post-mitotic neurons are generated in ventral r4 and pan-neuronal genes are thus expected to be found down-regulated in the mutants. We therefore favored for further analysis up-regulated over down-regulated genes. We chose six genes among the candidates that were overexpressed in the mutants for validation by ISH. One of them, Fgfr1, did not show changes noticeable by ISH (not shown). The others revealed three distinct patterns of changes. In the heterozygotes, Igfbp5 was expressed in only part of the Phox2b+ territory in the ventricular zone (VZ) and was excluded from the mantle layer (ML), while in the homozygotes, it showed a patchy distribution throughout the Phox2b+ VZ and was strongly expressed in the ML (Figure 2a, b). Igfbp5 is a member of a family of modulators of insulin-like growth factor (IGF) activity that may also have IGF-independent functions [13]. Like in our screen, Igfbp5 has obtained top scores in other differential screens on neural tissues (see, for instance, [14,15]), signifying perhaps that their expression is exceptionally labile. Their role in neural development has not been explored. Since Igfbp5 has been found to colocalize with areas of apoptosis during development [13], its overexpression may be related to the increased apoptosis caused by lack of Phox2b activity in ventral r4. Hes1, best known as an effector of Notch signaling [16,17], is normally weakly expressed in the floor plate and more strongly in the dorsalmost part of the Phox2b+ VZ in the E10.5 hindbrain, but it invades most of the normally Phox2b-expressing VZ in a salt-and-pepper pattern in the mutants (Figure 2c, d).
Figure 2.
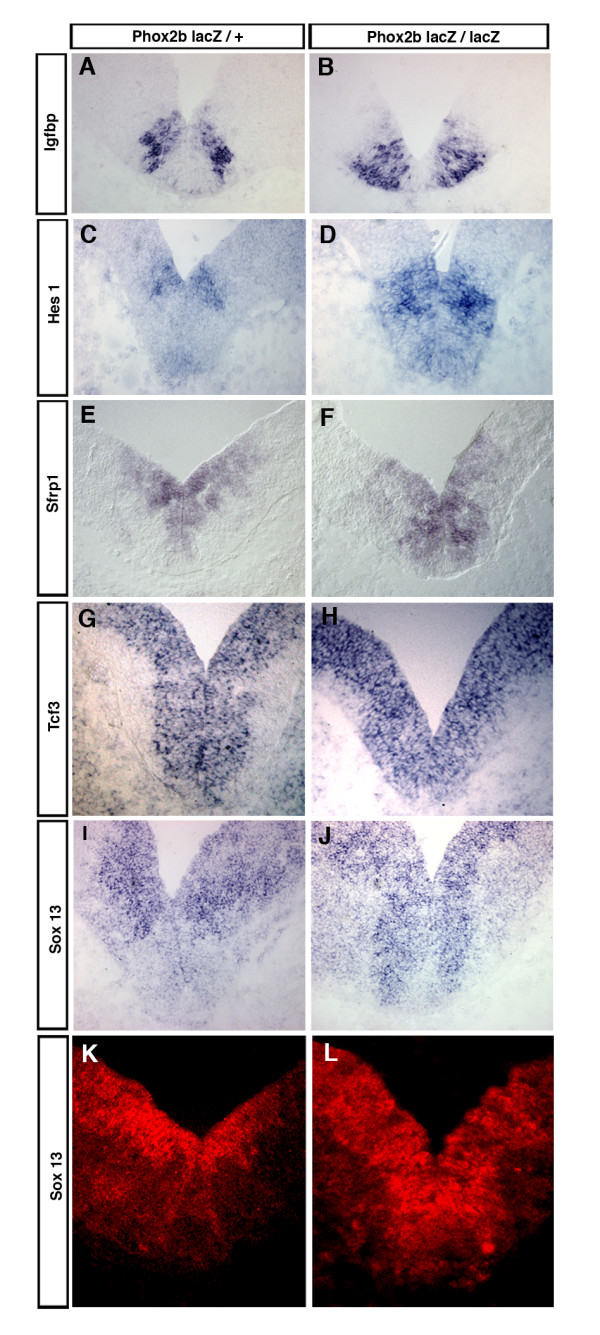
Expression patterns of a subset of genes up-regulated in ventral r4 of Phox2bLacZ/LacZ embryos. (a-j) In situ hybridizations on transverse sections of ventral r4 of Phox2bLacZ/+ (a, c, e, g, i) or Phox2bLacZ/LacZ (b, d, f, h, j) E10.5 mouse embryos. DIG-labeled riboprobes recognizing Igfbp5 (a, b), Hes1 (c, d), Sfrp1 (e, f), Tcf3 (g, h) and Sox13 (i, j) mRNA were used. (k, l) Immunofluorescence with an antibody recognizing Sox13 on transverse sections of ventral r4 of Phox2bLacZ/+(k) or Phox2bLacZ/LacZ (l) E10.5 mouse embryos.
We found Sfrp1 and Tcf3 to be normally expressed in and confined to the VZ, Sfrp1 only in the ventral part of the basal plate, and Tcf3 throughout the neural tube. In the mutant territory, their expression extended up to the neural tube border, thus including the post-mitotic neurons in the ML (Figure 2e–h). A similar expansion into the mutant ML has been found in previous work for Dll1 and Nkx2.2, which are normally confined to the VZ [4]. In contrast to Tcf3, Sfrp1 appeared also to be up-regulated throughout the mutant VZ: the signal given by the Sfrp1 probe was stronger outside of the fbm progenitors in Phox2bLacZ/+ embryos, but the reverse was true in Phox2bLacZ/LacZ animals. Tcfs are transcription factors that function in the Wnt signaling pathway [18,19]. Secreted frizzled-related proteins (Sfrps) also function mainly in the Wnt pathway as antagonists of Wnt signaling, but Sfrp1 may also exert a Wnt-independent activity as a stimulator of neurite outgrowth and as a growth cone attractant for retinal axons [20-22].
The Sox genes code for an extended family of SRY-box-containing transcriptional regulators [23]. Sox13, a member of the SoxD group, has been identified as a diabetes autoantigen expressed in pancreatic beta-cells [24] and recently as a positive regulator of gamma-delta T cells [25]. We found Sox13 to be expressed throughout most of the VZ, but almost absent in fbm progenitors. In the Phox2b mutants, its expression in the normally Phox2b+ VZ becomes as strong as elsewhere in the VZ (Figure 2i–l). We then asked whether Sox13 would also be excluded from the trigeminal bm progenitors in r2 that also express and depend on Phox2b. In ventral r2, Phox2b is expressed in the VZ and bm neuron production is underway at E9.5. One day later, bm neuron production has ceased and Phox2 is down-regulated in the VZ [7]. At E9.5, strong Sox13 expression in the VZ was interrupted at the border of the Phox2b+ territory, similar to what has been observed for the E10.5 fbm progenitors. At E10.5, when Phox2b expression was about to disappear in the VZ, Sox13 became clearly detectable in the ventralmost progenitor domain (Figure 3a–d). Cessation of Phox2b expression and bm neuron production in ventral r2 is accompanied by a fate switch of the progenitors, which now produce serotonergic neurons. Their production depends on Mash1 activity, in the absence of which serotonergic neurons are not produced [7]. We thus asked whether the expansion of Sox13 expression would take place in Mash1 mutants. Indeed, in Mash1-/- embryos, Sox13 expression did not invade the ventral progenitor domain, although Phox2b expression has ceased in the VZ (Figure 3e, f).
Figure 3.
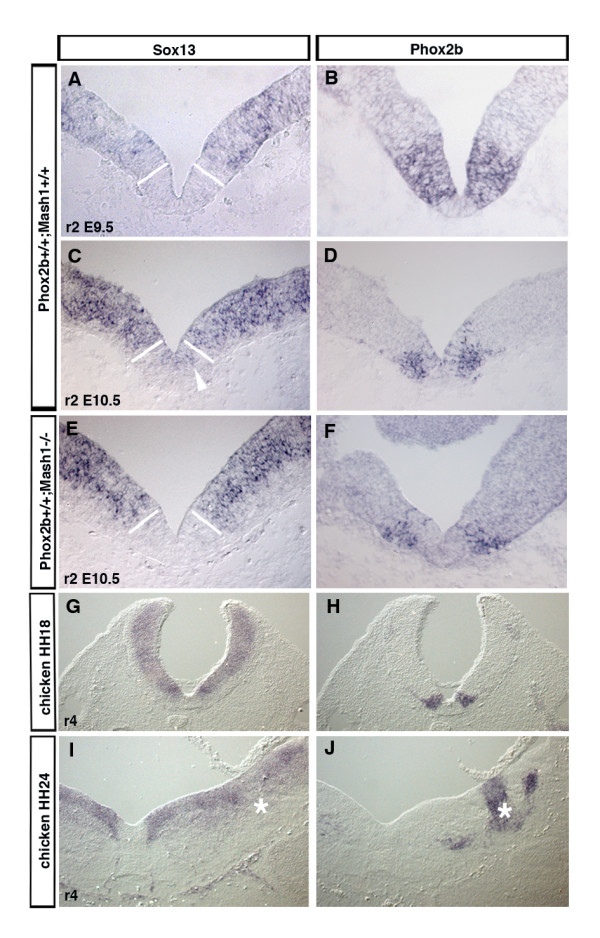
Sox13 is down-regulated by Phox2b-expressing cells in the hindbrain. (a-f) In situ hybridizations on transverse sections of ventral r2 from wild-type (a-d) or Mash1-/- (e, f) E9.5 (a, b) or E10.5 (c-f) mouse embryos with DIG-labeled riboprobes recognizing Sox13 (a, c, e) or Phox2b (b, d, f). At E9.5, Sox13 expression is excluded from the ventral progenitor domain expressing Phox2b, but invades the progenitor domain when Phox2b expression has disappeared at E10.5 in wild-type (arrowhead), but not in Mash1-/- embryos. The Phox2b-expressing region as visualized in (b, d, f) is delimited by white bars. (g-j) In situ hybridizations on transverse sections of ventral r4 of HH18 (g, h) or HH24 (i, j) chicken embryos with DIG-labeled riboprobes recognizing Sox13 (g, i) or Phox2b (h, j). Asterisks mark a lateral Phox2b+ progenitor domain, which does not express Sox13.
In Hamburger and Hamilton stage 18 (HH18) chicken hindbrain, we also found Sox13 expression very weak or absent in the Phox2b+ domain that gives rise to bm/visceromotor neurons, while it extended into the ventral progenitor domain at HH24, when Phox2b has disappeared there. At this stage, a distinct Phox2b+ progenitor domain has formed in the basal alar plate. Strikingly, Sox13 expression in the VZ was interrupted at this location (Figure 3g–j). Together, these results suggest that Phox2b represses Sox13 expression in the hindbrain VZ. The effect may be indirect since Sox13 continues to be down-regulated in the progenitors when Mash1 is absent.
Gap43, Sfrp1 and Sox13 are regulated by Phox2b misexpression in the spinal cord
We then selected Gap43, Sfrp1 and Sox13 for gain of function experiments in the chicken embryo spinal cord to ask whether they were regulated by ectopically expressed Phox2b. We chose Gap43 as a panneuronal Phox2b-dependent gene because it was more strongly expressed in fbm neurons, Sfrp1 as an example of the VZ genes that expand into the mutant ML since it was at the same time up-regulated in the VZ, and Sox13 because Phox2b repressed it in bm progenitors.
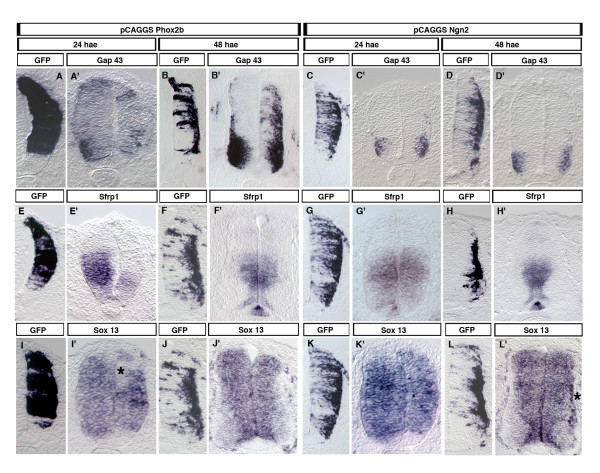
We thus examined whether Phox2b electroporated into the spinal cord of HH12-13 chicken embryos could up-regulate Gap43 and down-regulate Sfrp1 and Sox13 expression, in line with the changes observed in Phox2b homozygous mutants. Gap43, normally expressed at this stage mainly in the post-mitotic motoneurons, was weakly up-regulated in the VZ at 24 hours after electroporation (hae). It was massively induced in the ML at 48 hae (Figure 4a, b). To exclude the possibility that this was a trivial consequence of the fact that Phox2b triggered neuronal differentiation, we overexpressed Ngn2, which has a similar propensity to promote neuronal differentiation [6,26]. Ngn2, however, had no effect on Gap43 expression (Figure 4c, d). Sfrp1, normally expressed in a broad band of the VZ, was drastically down-regulated at 24 hae Not surprising for a cell-autonomous effect, Sfrp1 expression was only marginally affected at 48 hae, when the Phox2b-transfected cells had become post-mitotic and had moved to the ML. Ngn2 transfection did not induce changes in Sfrp1 expression (Figure 4e–h). The situation with regard to Sox13 was more complex. We found Sox13 expression to be confined to the VZ. At 24 hae in the Phox2b-transfected side, areas where Sox13 appeared to be repressed alternated with areas in which Sox13 was up-regulated, reflecting perhaps a highly dynamic expression pattern. Such a pattern was not seen after electroporation of Ngn2, which had instead a slight propensity of inducing Sox13 post-mitotically (Figure 4i–l). These results show that the changes in Gap43, Sfrp1 and Sox13 expression are not a simple consequence of the cells becoming prematurely post-mitotic and suggest that Phox2b does not exert its effect through a complex cascade of transcriptional events since the changes are already seen at 24 hae.
Figure 4.
Regulation of Gap43, Sfrp1 and Sox13 expression by Phox2b in the chicken spinal cord. (a-l') Chicken neural tubes electroporated at HH12-13 with pCAGGS::Phox2b-IRES-EGFP (a-b', e-f', i-j') or with pCAGGS::Ngn2 together with pCAGGS::EGFP (c-d', g-h', k-l') were analyzed 24 or 48 h after electroporation using DIG-labeled riboprobes recognizing GFP, Gap43, Sfrp1 or Sox13 mRNA. The asterisk in (i') marks cells in the transfected area where Sox13 was clearly down-regulated by Phox2b; it was rather up-regulated in adjacent cells. The asterisk in (l') marks cells in the transfected area where Sox13 was upregulated by Ngn2. Each experiment was repeated three times on different embryos with identical results.
GAP43 overexpression does not grossly affect neural tube morphology
Misexpression of Phox2b or its paralogue Phox2a in spinal regions of the neural tube induces neuronal differentiation and a bm neuronal fate and imposes an axonal phenotype that shares many features with that of bm neurons. Many dorsally located Phox2-transfected neurons, instead of growing commissural axons as they normally do, project into the periphery through dorsal exit points as bm neurons do, but also at ectopic sites [2] (Figure 5c, d). As Gap43 is up-regulated by Phox2b both in fbm precursors and when misexpressed in the spinal cord, we thus tested whether it would mimic this phenotype and, in particular, induce axonal growth when overexpressed in the chicken neural tube. However, after electroporation of Gap43, the morphology of the neuroepithelium remained grossly normal. Neither did the cells relocate to the ML nor were there any signs of aberrant or increased axonal growth as produced by Phox2b (Figure 5e, f).
Figure 5.
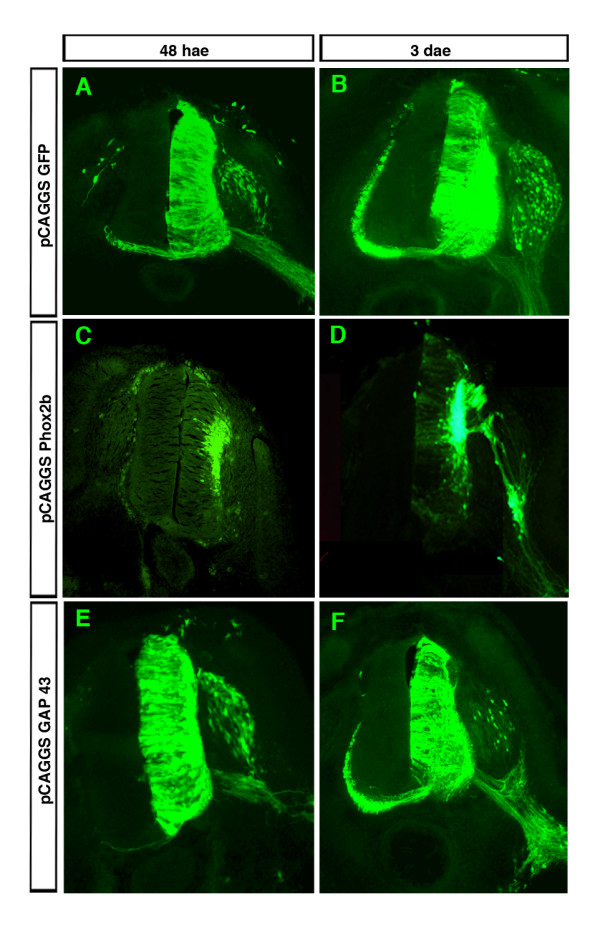
Overexpression of Gap43 does not mimic the effects of Phox2b on relocation to the mantel layer and axonal growth. (a-f) Vectors expressing either GFP alone (a, b) or Phox2b and GFP (c, d) or Gap43 and GFP (e, f) were electroporated into the spinal cord of HH12-13 chicken embryos and GFP was visualized by immunohistochemistry 48 hae (a, c, e) or 3 days after electroporation (dae) (b, d, f). Each experiment was repeated three times on different embryos with identical results.
Sfrp1 and Sox13 overexpression counteracts some of the effects of Phox2b misexpression
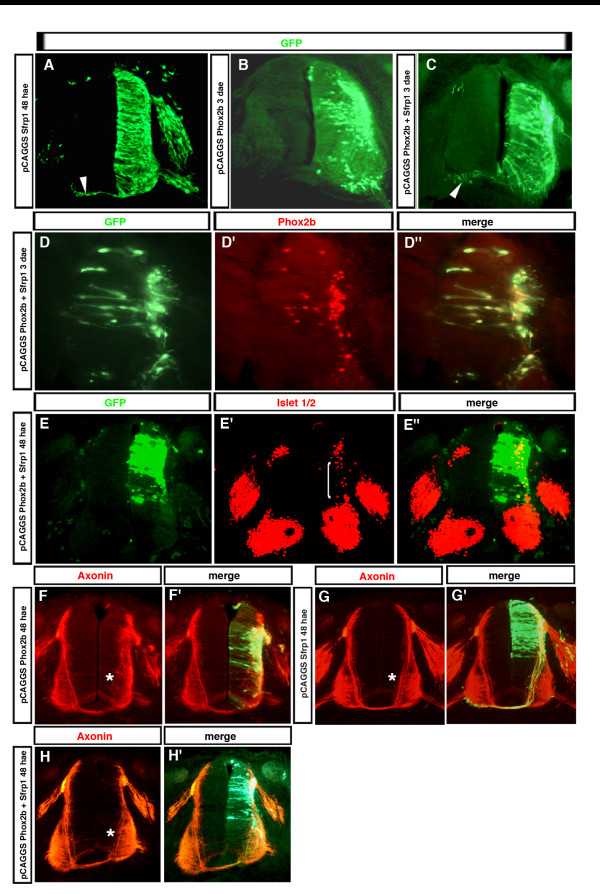
Overexpression of Sfrp1 in the chicken embryo spinal cord also did not produce gross anatomical alterations in the neural tube detectable by green fluorescent protein (GFP) labeling (Figure 6a, compare with Figure 5a). Since Sfrp1 appeared to be repressed by Phox2b in fbm precursors and when misexpressed in the spinal cord, we asked whether Sfrp1 would counteract some of the effects produced by Phox2b misexpression. After co-electroporation of Sfrp1 together with Phox2b, virtually all transfected cells expressed both GFP from the Sfrp1 expression vector and Phox2b, showing that co-expression is highly efficient (Figure 6d). When expressed together with Phox2b in this way, Sfrp1 did not prevent Phox2b from either driving neuronal differentiation and relocation to the ML or promoting ectopic neural tube exit of axons (Figure 6c, d; data not shown).
Figure 6.
Sfrp1 partially restores a commissural axonal phenotype. (a-c) Chicken neural tubes electroporated at HH12-13 with pCAGGS::Sfrp1-IRES-EGFP (a), pCAGGS::Phox2b-IRES-EGFP (b) or pCAGGS::Sfrp1-IRES-EGFP plus pCAGGS::Phox2b-IRES-EGFP (c) were analyzed by anti-GFP immunohistochemistry. Expression of Sfrp1 did not grossly alter the morphology of neuroepithelial cells, neither did it prevent Phox2b from promoting relocation to the ML. However, Sfrp1 prevented Phox2b from repressing the growth of commissural axons (arrowheads). (d-d") Chicken neural tubes electroporated with pCAGGS::Sfrp1-IRES-EGFP and a Phox2b expression vector were analyzed by anti-GFP and anti-Phox2b immunohistochemistry as indicated. Virtually all the transfected cells express both GFP and Phox2b. (e-e") Chicken neural tubes electroporated with pCAGGS::Sfrp1-IRES-EGFP plus pCAGGS::Phox2b-IRES-EGFP were analyzed by anti-GFP and anti-Islet1,2 immunohistochemistry as indicated. Co-expression of Sfrp1 did not prevent Phox2b from inducing Islet1,2 (bracket). (f-h') Chicken neural tubes electroporated with pCAGGS::Phox2b-IRES-EGFP (f, f'), pCAGGS::Sfrp1-IRES-EGFP (g, g') or with pCAGGS::Sfrp1-IRES-EGFP plus pCAGGS::Phox2b-IRES-EGFP (h, h') were analyzed by anti-axonin-1 and anti-GFP immunohistochemistry. In (f, g, h), the anti-axonin-1 staining is shown alone, and in (f', g', h') it is merged with the anti-GFP immunofluorescence. The fascicle formed by the commissural fibers en route to the floor plate is marked by an asterisk; it is absent after Phox2b transfection and partially restored by co-expressing Sfrp1.
However, forced expression of the Phox2 genes [2] (Figures 5c and 6b) suppresses the growth of commissural axons, while cells co-transfected with Sfrp1 and Phox2b clearly grew commissural fibers as revealed by GFP staining in five out of five electroporated embryos (that is, n = 5/5; Figure 6c). To visualize the trajectory of pre-crossing commissural axons in electroporated embryos, we used anti-axonin-1 (TAG-1 in mammals) antibodies known to strongly label commissural fibers [27]. Sfrp1 overexpression did not change the normal pattern of extension of commissural axons, which form a conspicuous fascicle running along the medial border of the motoneuron column (Figure 6g; n = 4/4). As expected, the axonin-1-expressing fascicle was absent at the transfected side after Phox2b electroporation (Figure 6f; n = 9/9). Still, anti-axonin-1-labeled fibers were observed in the Phox2b-transfected dorsal spinal cord, but they neither formed a tight fascicle nor extended beyond the dorsal border of the motoneuron column. Co-expressing Sfrp1 together with Phox2b partially, but not completely, restored the axonin-1-labeled fascicle that extends towards the floor plate (Figure 6h; n = 6/6).
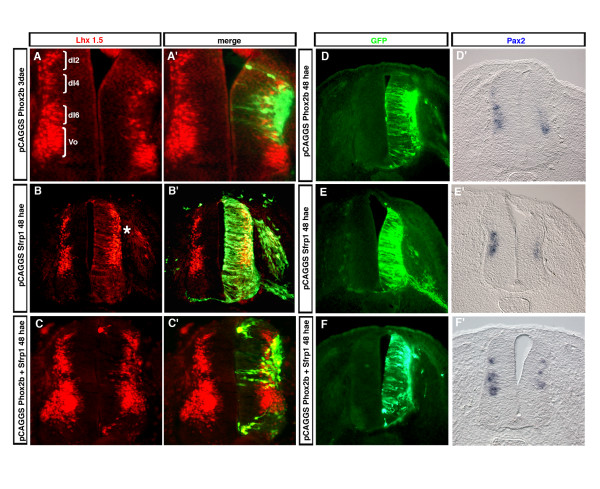
The commissural axonal phenotype prompted us to ask whether Sfrp1 may counteract some of the fate changes affected by Phox2b. Sustained expression of Sfrp1 together with Phox2b reduced, but did not abrogate, the induction of Islet1,2 (Figure 6e; n = 3/3), normally seen after transfecting Phox2b alone [2,6]. Phox2b, in addition to inducing bm markers, represses the interneuronal marker Lhx1,5 [2] (Figure 7a), which is normally expressed by the interneuron populations dI2, dI4, dI6 and V0 [28,29]. This effect was offset by co-expressing Sfrp1 (Figure 7c; n = 5/5). Hence, the cells now expressed a mixed phenotype, which is not normally encountered: Phox2b+, Islet1, 2+, Lhx1,5+. Sfrp1 increased the number of Lhx1,5-expressing cells in the dorsal spinal cord also when expressed alone, explaining its ability to counteract the down-regulation of Lhx1,5 caused by Phox2b. After Sfrp1 electroporation, the gap in Lhx1,5 expression corresponding to the dI5 neurons was not observed anymore at the transfected side (Figure 7b; n = 9/10). A fainter but clearly visible signal was also seen in the Sfrp1-transfected VZ in cells with neuroepithelial morphology, whereas it was never observed in control conditions, suggesting that Lhx1,5 is induced ectopically in the presence of Sfrp1 (Figure 7b; n = 9/10). Finally, we examined Pax2, which is expressed by several Lhx1,5+ populations [28], as a second marker of spinal interneurons. As expected, misexpression of Phox2b also down-regulated Pax2 in the transfected cells (Figure 7d; n = 3/3). In contrast to Lhx1,5, however, Pax2 was down-regulated as well by overexpressing Sfrp1 alone (Figure 7e; n = 3/3), and consequently, Sfrp1 did not prevent Phox2b from down-regulating Pax2 (Figure 7f; n = 3/3). Hence, Sfrp1 overexpression affects the phenotype of the neurons that are generated and thwarts the ability of Phox2b to induce a bm phenotype.
Figure 7.
Changes in Lhx1,5 and Pax2 expression after Sfrp1 transfection. (a-c') Chicken neural tubes electroporated at HH12-13 with pCAGGS::Phox2b-IRES-EGFP (a, a'), pCAGGS::Sfrp1-IRES-EGFP (b, b') or with pCAGGS::Sfrp1-IRES-EGFP plus pCAGGS::Phox2b-IRES-EGFP (c, c') were analyzed by anti-Lhx1,5 and anti-GFP immunohistochemistry. In (a, b, c), the anti-Lhx1,5 staining is shown alone, and in (a', b', c') it is merged with the anti-GFP immunofluorescence. Phox2b represses Lhx1,5 normally expressed in interneuron populations dI2, dI4, dI6 and V0 (brackets), while Sfrp1 increases the number of Lhx1,5+ cells (asterisk) and counteracts Phox2b in its capacity of repressing Lhx1,5 (compare (a) and (c)). (d-f') Chicken neural tubes electroporated at HH12-13 with pCAGGS::Phox2b-IRES-EGFP (d, d'), pCAGGS::Sfrp1-IRES-EGFP (e, e') or with pCAGGS::Sfrp1-IRES-EGFP plus pCAGGS::Phox2b-IRES-EGFP (f, f') were analyzed by anti-GFP immunohistochemistry or by ISH with a Pax2 probe as indicated. Pax2 is down-regulated in all three conditions.
Sfrp1 (SARP2) has been found to sensitize cells in culture to pro-apoptotic stimuli, whereas Sfrp2 had the opposite effect [30] and prevented programmed cell death in the neural crest [31]. We thus tested whether Sfrp1 might influence cell death in our overexpression paradigm. In line with published data [32], we found very few apoptotic cells in the neural tube at the stages studied (one to four TUNEL+ cells per section), and their number differed neither between electroporated and non-electroporated sides of the neural tube nor among different conditions (electroporation of Srp1 or Phox2b or of Sfrp1 plus Phox2b) (not shown).
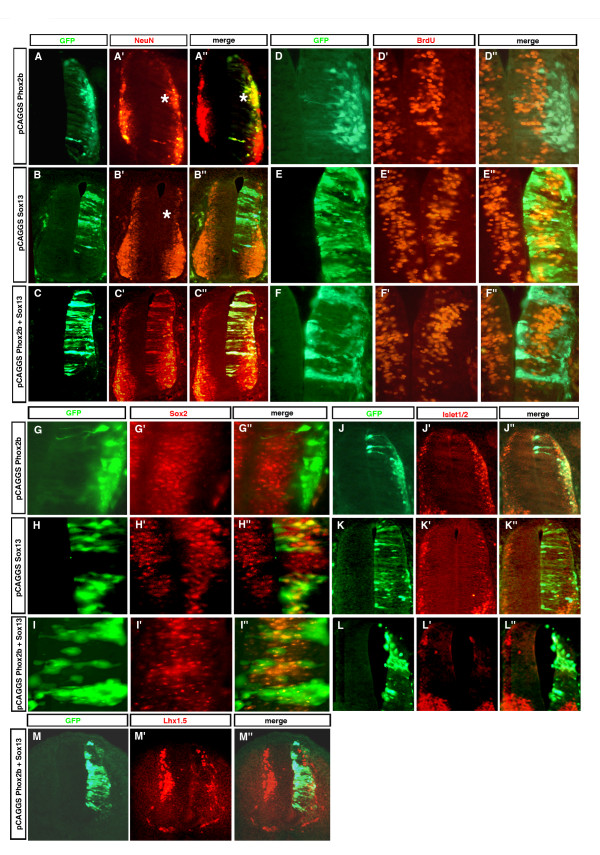
In the chicken and mouse VZ, cells highly expressing Sox13 alternate with cells that express it less (Figures 2i, k and 4i', j'), a pattern that seems to be accentuated by overexpressing Phox2b (Figure 4i'). Such a pattern suggests that Sox13 expression in neuroepithelial cells is dynamic, perhaps in relation to their commitment to a neuronal fate. VZ expression of Sox13 driven by a strong constitutive promoter should cancel out these changes in expression. When overexpressed in this way, Sox13 did not disrupt neural tube structure or grossly alter neuroepithelial cell morphology. The transfected cells incorporated bromo-deoxyuridine (BrdU; Figure 8e; n = 3/3) and most expressed the VZ cell marker Sox2 (Figure 8h; n = 3/3). However, there were less cells expressing the neuronal marker NeuN in the transfected area, and none of the transfected cells co-expressed NeuN (Figure 8b; n = 3/3), indicating that constitutively high expression of Sox13 inhibits neuronal differentiation.
Figure 8.
Sox13 overexpression partly antagonizes Phox2b activity. (a-m") Chicken neural tubes electroporated at HH 12–13 either separately with pCAGGS::Phox2b-IRES-EGFP (a-a", d-d", g-g", j-j") or pCAGGS::Sox13-IRES-EGFP (b-b", e-e", h-h", k-k") or with pCAGGS::Phox2b-IRES-EGFP plus pCAGGS::Sox13-IRES-EGFP (c-c", f-f", i-i", l-l", m-m") were analyzed 48 h after electroporation. Transverse sections were stained with anti-GFP antibodies in combination with anti-NeuN (a-c"), anti-Sox2 (g-i"), anti-Islet1/2 (j-l") or anti-Lhx1,5 immunohistochemistry (m-m") or in combination with anti-BrdU antibodies (d-f") after BrdU injection into the amniotic cavity 2 h before fixation. Phox2b induces and Sox13 represses NeuN (asterisks in (a', a", b')) in the ML. When co-expressed with Sox13, Phox2b is still capable of inducing NeuN, which, however, is now also switched on in the VZ (c', c"). Cells electroporated with Phox2b are BrdU-negative whether Sox13 was co-transfected (f) or not (d). Cells electroporated with Sox13 whether co-transfected with Phox2b (i) or not (h) express Sox2, while cells electroporated with Phox2b alone are always Sox2-negative (g). Islet1,2 induction by Phox2b (j) is abolished by co-transfection of Sox13 (l). Repression of Lhx1,5 by Phox2b is not prevented by co-expressing Sox13 (m). Co-transfection of Sox13 together with Phox2b inhibits relocation to the ML (c, f, i, l, m) implemented by expression of Phox2b alone (a, d, g, j).
We next tested the idea that Sox13 overexpression may antagonize Phox2b in its capacity to drive neuronal differentiation. Phox2b misexpressed alone causes the transfected cells to accumulate in the ML and to express NeuN (Figure 8a), as expected from its propensity to promote neuronal differentiation and relocation to the ML. When Sox13 was misexpressed together with Phox2b, many transfected cells remained in the VZ where they either retained the morphology of neuroepithelial cells or aggregated near the lumen of the neural tube (Figure 8c, f, l; n = 3/3). Phox2b was still able to induce NeuN, but now many cells double-positive for NeuN and GFP were located in the VZ and had the morphology of neuroepithelial cells. Hence, Sox13, although unable to inhibit induction of NeuN by Phox2b, appears to prevent the transfected cells from losing their neuroepithelial morphology and from relocating to the ML. We then investigated further the phenotype of the cells constitutively co-expressing Phox2b and Sox13. We first established that, in the presence of Sox13, Phox2b was still able to promote cell cycle exit as shown by the fact that the transfected cells did not incorporate BrdU (Figure 8f; n = 3/3). However, while the neural stem cell marker Sox2 [33] was never expressed in cells transfected with Phox2b alone, many doubly transfected cells still expressed Sox2 (Figure 8g, i; n = 3/3). Finally, we examined Islet1,2 as a marker for the bm phenotype induced by Phox2b, and Lhx1,5 as a marker for interneurons. Induction of Islet1,2 was completely repressed by co-transfection with Sox13 (Figure 8j, l; n = 3/3). By contrast, Sox13 did not prevent down-regulation of Lhx1,5 after Phox2b transfection, a result to be expected since Sox13 overexpression inhibits neuronal differentiation (Figure 8m; n = 3/3).
Discussion
Phox2b plays a key role in the formation of two important classes of cranial motoneurons, the bm and the closely related visceromotor neurons. Its role in motoneuronal differentiation has been studied in most detail during differentiation of the fbm neurons. This work has yielded a few genetic targets whose expression is dependent on Phox2b in fbm precursors [4,10]. To search for Phox2b-regulated genes by a more systematic approach, we have used gene expression profiling of ventral r4, in which the fbm precursors arise, from heterozygous and homozygous Phox2b mutant embryos. In this way we have identified a number of candidates that are either up- or down-regulated in the homozygous mutants. Two of these candidates regulated by Phox2b had been identified in previous work, but none of the others have been reported before. We restrict our discussion to those candidates we confirmed by ISH as being regulated by Phox2b in ventral r4.
Our analysis did not distinguish between genes that were under direct control of Phox2b and genes that depended on Phox2b indirectly, through the action of other transcription factors. However, the genes that we found induced or repressed in the chicken spinal cord already at 24 hae can be supposed to be proximal targets in the genetic program induced by Phox2b, and not to be dependent on a vast network of other factors. So far, only transcriptional activation by Phox2b has been demonstrated, but the possibility exists that it may also act as a repressor, as indicated by the observation that repressor forms of Phox2b reproduced some of the effects of Phox2b [6].
Since less post-mitotic neurons are produced in the mutant territory, it was to be expected that many genes that are normally expressed in young post-mitotic neurons would be underrepresented in Phox2bLacZ/LacZ embryos. App, Mapk8, Nfl and Gap43 belong to this category. Although post-mitotic cells are produced by Phox2bLacZ/LacZ progenitors and migrate to the ML, albeit in reduced numbers, the ML cells failed to express these genes. The same pattern has been found in previous work for other post-mitotic markers [4]. Some of the post-mitotic cells that arise in the mutants then go on to become serotonergic neurons [1,7]. Apparently, in the absence of Phox2b and its proneural activity, these cells will acquire neuronal properties only at later stages. Gap43 is a case apart since its expression in post-mitotic neurons was distinctly stronger in fbm precursors than elsewhere in r4, suggesting that high Gap43 activity may be required for proper fbm differentiation. Gap43 was also markedly up-regulated by ectopic Phox2b expression in the spinal cord, an effect that could not be ascribed to an increase in generic neuronal differentiation since Ngn2 overexpression was without effect. Gap43 was originally described as a protein associated with growing axons, both in development and during regeneration, and is thought to function as a determinant of axonal growth competence [11,12,34]. Despite a wealth of in vitro data describing its growth-promoting properties, its in vivo function is still ill-defined. When constitutively expressed in adult mice, GAP43 induced branching of motor nerve terminals and promoted sprouting of nerves after lesion [35]. In our study, Gap43 overexpression in the embryonic neural tube did not produce overt morphological alterations. Hence, its effects might be limited to terminal fields and situations of regeneration. A number of discrete developmental defects have been reported in Gap43 mutants, including pathfinding defects of the optic nerve [36], a reduction in serotonergic innervation of the forebrain [37] and a failure to form the anterior commissure [38]. Such a subtle phenotype for a gene expressed in virtually all young neurons may be due to redundancy with the CAP23 gene [39].
Among the genes found to be up-regulated in mutant ventral r4, Hes1, at the stage studied, was expressed only in the dorsalmost part of the Phox2b+ VZ in wild-type or heterozygous animals. In Phox2bLacZ/LacZ embryos, its expression invaded most of the normally Phox2b+ domain. The basic-helix-loop-helix transcription factor of the hairy and enhancer of split family, Hes1 is best known as an effector of Notch signaling and, together with its paralogues Hes3 and Hes5, plays a crucial role in maintaining the progenitor state of neuroepithelial cells [16,17]. Since Phox2b drives neural progenitors to exit the cell cycle and to differentiate, it is perhaps not surprising that it shuts off a negative regulator of neural differentiation. This scheme, however, is complicated by the observation that its paralogue Hes5 has been found to be down-regulated in Phox2bLacZ/LacZ embryos [8]. It appears that the lack of proneural activity in Phox2b mutants and the consecutive down-regulation of the Notch ligand Dll1 in the VZ affect differently the two paralogous genes. The Lef/Tcf proteins are transcription factors that play pivotal roles in Wnt signaling, functioning mainly as repressors in the absence and as obligatory co-activators of beta-catenin in the presence of Wnt ligands [19]. Tcf3 has remained the most enigmatic member of the family. Loss of Tcf3 results in early gastrulation defects and perhaps as a consequence to broad defects in neural tube patterning [40]. Tcf3 has been said to become undetectable in the mouse embryo from E10.5 onward [18], but we found it strongly expressed in E10.5 r4 throughout the VZ. Normally excluded from the ML, its expression domain invaded the ML in the mutant territory. This has been observed in previous work also for Nkx2.2 and Dll1 [41]. Apparently, the post-mitotic cells that accumulate in the mutant ML are not only unable to express generic neuronal markers but also do not switch off genes whose expression is normally confined to the VZ.
Sfrp1 shared with Tcf3 the tendency to be expressed in the mutant ML, from which it was normally excluded. However, the expression pattern of Sfrp1 in r4 was much more circumscribed than that of Tcf3. It was only expressed in ventral r4, including the Phox2b+ progenitor domain and extending up to the middle of the basal plate. In Phox2bLacZ/+ embryos, its expression was stronger dorsal to the Phox2b+ progenitors, but the reverse was true in Phox2bLacZ/LacZ embryos. In the embryonic chicken spinal cord, we found Sfrp1 to be strongly expressed in the VZ of the dorsal basal plate, in line with previous results [42], and to be massively repressed by Phox2b. Sfrp1 overexpression in the chicken spinal cord up-regulated the interneuron marker Lhx1,5. When co-expressed with Sfrp1, Phox2b was still capable of inducing Islet1,2, but became unable to repress expression of Lhx1,5. Hence, co-expression of Phox2b and Sfrp1 results in a mixed phenotype that is not normally encountered in the spinal cord, as also indicated by the observation that Sfrp1 partially restored the ability of Phox2b-transfected neurons to grow commissural axons. However, expression of Pax2, another interneuronal marker that was down-regulated by Phox2b, was already reduced by expression of Sfrp1 alone and continued to be down-regulated when Sfrp1 was co-expressed with Phox2b.
The Sfrps have been mostly described as secreted inhibitors of Wnt signaling, but there is evidence that they are able to promote rather than to inhibit Wnt signaling in some contexts and that they can act through Wnt-independent mechanisms [20-22]. In the Wnt1-/-; Wnt3a-/- embryonic spinal cord, the dI4 (Lhx1,5+; Pax2+) subclass of interneurons has been found to be expanded at the expense of the dI1 and dI2 interneuron populations [43]. In line with these results, we found that Sfrp1 increased the population of Lhx1,5-expressing cells. Sfrp1, however, down-regulated Pax2, suggesting that the effect of Sfrp1 overexpression cannot be explained solely by inhibition of Wnt signaling. Gain of function experiments in the chicken retina have shown that Sfrp1 promotes neuronal differentiation and the generation of retinal ganglion cells and cones at the expense of other cell types by a mechanism that does not involve inhibition of Wnt signaling [44]. In our study, Sfrp1 overexpression in the spinal cord did not cause an obvious increase in generic neuronal differentiation. Still, our results suggest that Sfrp1 plays a role in neuronal subtype specification as has been observed in the retina.
Transcription factors of the B-, C- and E-group Sox proteins have been found to play key roles during early neural development [33,45-47]. By contrast, much less is known about the expression of the D-group Sox proteins in neural tissues and virtually nothing is known about their function during central nervous system development. In our screen, the D-group Sox13 gene was the only Sox gene that scored as a Phox2b-regulated gene. A previous study reported Sox13 to be expressed mainly by differentiating neurons in the ML [48]. At the slightly earlier stages and more caudal regions studied here, we found, by both ISH and antibody staining, Sox13 to be expressed throughout and confined to the VZ in both the mouse and chicken nervous systems. We found Sox13 to be expressed in the VZ in a salt-and-pepper pattern, patches of high signal intensity alternating with cells expressing lower levels, suggesting a highly dynamic expression pattern perhaps in relation to the commitment to differentiation. VZ cells that expressed constitutively high levels of Sox13 after transfection were proliferating as indicated by BrdU incorporation and many expressed the neural cell stem marker Sox2, but none the neuronal marker NeuN, whose expression in the ML was inhibited. These results may be interpreted to mean that the cells normally expressing lower levels were those destined to become neurons. In E10.5 ventral r4, Sox13 mRNA and protein expression, which was strong in more dorsal regions, was much weaker in the fbm progenitor domain, but invaded the normally Phoxb+ region in the mutants. While Phox2b expression in the VZ and motor neuron production is prolonged in ventral r4, peak motor neuron production in r2 occurs at E9.5 and ceases around E10.5 together with Phox2b expression in the VZ, where the progenitors switch to the production of serotonergic neurons [1]. Consequently, in E9.5 ventral r2, Sox13 expression was absent from ventral progenitors, but became detectable there one day later. At this stage, the basic-helix-loop-helix factor Mash1 has been found to be required in this domain as both the sole provider of proneural activity and a determinant of the serotonergic phenotype [7]. As shown here, Sox13 expression did not expand into the ventral progenitor domain when Mash1 was absent, although Phox2b expression had ceased. One possibility is that Sox13 expression requires activation of the Notch pathway by its ligands, whose expression depends on proneural activity provided by Mash1, but more work is required to ascertain this. The idea that Sox13 may function as a Notch effector is also supported by the observation that its overexpression inhibits neurogenesis.
In contrast to what might have been expected from the pronounced down-regulation of Sox13 in Phox2b-expressing progenitors, Phox2b misexpression in spinal regions of the chicken neural tube did not result in downright repression, but areas of down-regulation alternated with regions with enhanced expression. Again, one reason may be that Sox13 expression depends on Notch activity, which may be repressed by Phox2b but enhanced in neighboring cells by the propensity of Phox2b to induce the Notch ligand Dll1 [8]. Constitutively high expression of Sox13, however, counteracted Phox2b activity in our ectopic expression paradigm. A major effect was to block the relocation of the transfected cells to the ML. In the presence of forced expression of Sox13, Phox2b was still able to promote cell cycle exit and neuronal differentiation, but now the Phox2b+, NeuN+ and BrdU- cells normally located in the ML accumulated in the VZ. Many of the doubly transfected cells expressed Sox2, which was totally absent from cells transfected with Phox2b alone, and Islet1,2 induction by Phox2b was inhibited in them. Hence, Sox13 appears to counteract the capacity of Phox2b to silence Sox2 expression and to induce a bm phenotype. However, blocking migration to the ML may still be the primary effect, since persistent Sox2 expression and inhibition of Islet1,2 induction may be consequences of the former. So far, the only in vivo function of Sox13 that has been reported is the promotion of, and a requirement for, proper development of the gamma/delta T cell lineage [25]. This has been attributed to the inhibition of Wnt signaling by direct interaction with the Wnt effector TCF1. Given that this also holds true for Sox13 function in the nervous system, it is interesting that two inhibitors of Wnt signaling, Sfrp1 and Sox13, appeared in our screen. However, Sfrp1 and Sox13 act on different processes: Sox13 inhibits generic neuronal differentiation while Sfrp1 affects cell type specification. Repression of both may thus be a prerequisite for proper development of bm neurons. Sox13 knockout mice die at birth from unknown reasons [25], and this may well be because of defective neural development. Our results showing that Sox13 inhibits neurogenesis and blocks migration to the ML give a first glance of the possible in vivo functions of Sox13 in neural development, opening the way for future mechanistic studies.
Materials and methods
Mice and embryo dissection
Phox2bLacZ/+ mice [49] were intercrossed and the hindbrains of E10.5 embryos were dissected out and incubated for 2 minutes at 37°C with 1 μg/μl fluorescein di-(beta-D-galactopyranoside) (Sigma-Aldrich, Lyon, France). The left and right beta-galactosidase expressing domains of ventral r4 with the floorplate in between were excised under a fluorescent binocular and stored at -20°C in RNALater solution (Ambion, Austin, TX, USA). The rest of the embryo was used for genotyping as described by Pattyn et al. [49]. Mash1+/- mice (obtained from François Guillemot) were intercrossed and the embryos genotyped as described [50].
RNA extraction and amplification
Four r4 samples of the same genotype (either Phox2bLacZ/+ or Phox2bLacZ/LacZ) were pooled together and RNA was extracted and amplified using the RNAqueous-Micro Kit (Ambion) and the Message Amp aRNA amplification kit (Ambion) according to the instructions of the manufacturer. The quantity and the quality of the amplified RNA were verified using Agilent RNA 6000 Nano Assay and Bioanalyzer 2100 (Agilent Technologies, Massy, France).
cDNA microarray hybridization
Microarray hybridization was done using two cDNA collections, the NIA Mouse 15 k clone sets [51] and the NeuroDev clone sets (Ecole normale supérieure local collection), both spotted on Ultra GAPS II slides (Corning, Avon, France) using a BioRobotics MicroGrid microarrayer (Genomic Solutions, Huntingdon, UK) by the Transcriptome platform of ENS, Paris. Amplified RNA (2 μg) was retrotranscribed and labeled with dUTP-Cy5 or dUTP-Cy3 (Amersham, Piscataway, NJ, USA) using Superscript III Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) in the presence of 5 μg of random primers and appropriate buffer (Invitrogen). RNA was degraded by alkaline lysis and cDNA was purified using a QIAquick purification kit (Qiagen, Courtaboeuf, France). CDNA microarrays were pre-hybridized for 1 h at 42°C in 5 × SSC, 0.1% SDS, 1% bovine serum albumin, washed with distilled water and dried. They were hybridized overnight at 42°C in 25% formamide, 5 × SSC, 0.1% SDS with Cy5- and Cy3-labeled cDNA, washed in 1 × SSC, 0.2% SDS at 42°C, in 0.1 × SSC, 0.2% SDS and two times in 0.1 × SSC at room temperature and then spin-dried. Four biological replicas (independent dissections) for each microarray (15 k and NeuroDev) were done and each of these replicas was hybridized independently twice by inverting the labeling dyes (dye-swap). A total of 16 hybridizations where thus performed, 8 using NIA 15 k microarrays and 8 others using NeuroDev slides.
Data acquisition and analysis
The hybridized microarrays were scanned using Genepix 4000B (Molecular Devices, Sunnyvale, CA, USA)) and the resulting image files analyzed by GenePix Pro 5.0 software (Axon). For each GenePix output file, two filters were applied, one to clear out spots and another one to discard saturating spots where the median foreground intensity was greater than 60,000 in one of the two channels. The resulting median foreground intensities were normalized, without background signal subtraction, for dye bias using a global Lowess correction [52]. An expression matrix for each type of microarray (eight experiments each) was created gathering all normalized ratio data (M values). After normalization, we applied on each matrix a hierarchical clustering algorithm [53] using TMeV software [54] to detect a possible bias introduced by sample heterogeneity. The cluster obtained showed a clear separation according to dye-swap between hybridizations indicative of homogenous samples. The two matrices obtained were processed using the GEPAS web server [55] to remove genes with more than 30% missing values. A KNN impute step was performed to calculate missing values for the remaining genes using the 15 nearest neighbor profiles. A further scaling step was performed by dividing the ratios for each gene in one experiment by the experimental standard deviation and multiplying each value by the mean of all experimental standard deviations. The ratios were displayed such that a positive ratio describes a gene whose expression is higher in Phox2bLacZ/+ than in Phox2bLacZ/LacZ embryos. SAM [56] was used to search for differentially expressed genes and to take advantage of our four biological replicates to calculate the SAM score equivalent to the T-statistic value and to estimate the associated false discovery rate. We fixed the false discovery rate at 5% using the 90th percentile of falsely called genes. We chose in addition a SAM score above 3.5 or below -3.5 (for genes down- or up-regulated in the mutants, respectively) as an arbitrary cut-off since smaller scores increased the number of genes whose expression changes could not be confirmed by ISH.
Expression vectors and electroporation
We used pCAGGS::Phox2b-IRES2-EGFP [2], pCAGGS::Gap43-IRES2-EGFP, pCAGGS::Sox13-IRES2-EGFP, pCAGGS::Sfrp1-IRES2-EGFP or pCAGGS::IRES2-EGFP to express mouse Phox2b, mouse Gap43, human SFRP1 (the latter two subcloned from IMAGE clones) and human Sox13 (subcloned from Ultimate ORF Clone IOH44934, Invitrogen) together with GFP or GFP alone in the chicken neural tube driven by a composite chicken beta actin-CMV promoter. The spinal region of HH12-14 embryonic chick neural tubes were electroporated in ovo with the appropriate expression vectors (2 mg/ml or 1 mg/ml) essentially as described [8]. Correct expression of all constructs was verified by ISH with the cognate probes. After electroporation, embryos were allowed to develop at 38°C for 24–72 h.
Histological methods
Mouse embryos and well-electroporated chicken embryos, as assessed by GFP fluorescence, were dissected out, fixed in 4% paraformaldehyde overnight, embedded in Tissue-Tek® OCT™ (Sakura-Finetek, Zoeterwoude, The Netherlands) (mouse embryos) or gelatin (chicken embryos) and analyzed on 12 or 20 μm transverse cryosections, respectively. The methods used for ISH immunohistochemistry were as described previously [2,8]. The following antibodies were used: rabbit anti-axonin-1 [27], mouse monoclonal anti-BrdU (1/100; Sigma), mouse monoclonal (Roche, Basel, Switzerland) and rabbit (Chemicon, Temecula, CA, USA) anti-GFP (1/400), mouse anti-Islet1,2 (1/100) [57], rabbit anti-Islet1 (1/500; Abcam, Paris, France), mouse anti-Lhx1,5 (Developmental Studies Hybridoma Bank), mouse anti-NeuN (1/500), rabbit anti-Sox2 (1/1,000; Abcam), rabbit anti-Sox13 [48] and adequate fluorescent secondary antibodies. For NeuN and Sox2 staining, sections were pre-treated with sodium citrate pH 6, 0.05% Tween-20, and for BrdU staining, sections were pretreated in 2N HCl, 0.5% Triton X-100 for 20 minutes at room temperature and neutralized. Apoptotic cells were detected using the Apo Tag kit (Oncor, Gaithersburg, MD, USA)) according to the manufacturer's instructions.
Abbreviations
bm: branchiomotor; BrdU: bromo-deoxyuridine; E: embryonic day; fbm: facial bm; GFP: green fluorescent protein; hae: hours after electroporation; HH: Hamburger and Hamilton stage; IGF: insulin-like growth factor; ISH: in situ hybridization; ML: mantle layer; r: rhombomere; SAM: significance analysis of microarray; VZ: ventricular zone.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
PP devised and performed experiments and analyzed data, MRH performed experiments, SLC analyzed data, SR performed experiments, VH provided valuable reagents, CG devised research, analyzed data and wrote the article. All authors read and approved the final manuscript.
Supplementary Material
Genes down-regulated in Phox2bLacZ/LacZ embryos. A list of the 51 genes that were found to be significantly down-regulated in the ventral r4 of Phox2bLacZ/LacZ embryos according to the criteria presented in Materials and methods (SAM score above 3.5). The expression of the genes written in red was tested by ISH in Phox2bLacZ/+ and Phox2bLacZ/LacZembryos.
Genes up-regulated in Phox2bLacZ/LacZ embryos. A list of the 23 genes that were found to be significantly up-regulated in the ventral r4 of Phox2bLacZ/LacZ embryos according to the criteria presented in Materials and methods (SAM score below -3.5). The expression of the genes written in red was tested by ISH in Phox2bLacZ/+ and Phox2bLacZ/LacZembryos.
Acknowledgments
Acknowledgements
We like to thank F Guillemot for providing Mash1 mutant mice, ET Stoeckli for providing us with anti-axonin-1 antibodies, P Bovolenta for the chicken Sfrp1 probe and E Matsunaga for the chicken Pax2 probe. The monoclonal anti-Lhx1,5 antibodies developed by T Jessell were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biological Sciences. This work was supported by grants from Agence Nationale de la Recherche, Association Française contre les Myopathies, Association pour la Recherche sur le Cancer and the Fondation Franco-Norvegienne pour la Recherche Scientifique et Technique.
Contributor Information
Patrick Pla, Email: Patrick.Pla@curie.u-psud.fr.
Marie-Rose Hirsch, Email: hirsch@biologie.ens.fr.
Stéphane Le Crom, Email: lecrom@biologie.ens.fr.
Simone Reiprich, Email: enomis83@gmx.net.
Vincent R Harley, Email: Vincent.Harley@princehenrys.org.
Christo Goridis, Email: goridis@biologie.ens.fr.
References
- Pattyn A, Vallstedt A, Dias JM, Samad OA, Krumlauf R, Rijli FM, Brunet JF, Ericson J. Coordinated temporal and spatial control of motor neuron and serotonergic neuron generation from a common pool of CNS progenitors. Genes Dev. 2003;17:729–737. doi: 10.1101/gad.255803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch MR, Glover JC, Dufour HD, Brunet JF, Goridis C. Forced expression of Phox2 homeodomain transcription factors induces a branchio-visceromotor axonal phenotype. Dev Biol. 2007;303:687–702. doi: 10.1016/j.ydbio.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Sussel L, Serup P, Hartigan-O'Connor D, Jessell TM, Rubenstein JL, Ericson J. Homeobox gene Nkx2.2 and specification of neuronal identity by graded Sonic hedgehog signalling. Nature. 1999;398:622–627. doi: 10.1038/19315. [DOI] [PubMed] [Google Scholar]
- Pattyn A, Hirsch MR, Goridis C, Brunet JF. Control of hindbrain motor neuron differentiation by the homeobox gene Phox2b. Development. 2000;127:1349–1358. doi: 10.1242/dev.127.7.1349. [DOI] [PubMed] [Google Scholar]
- Ohsawa R, Ohtsuka T, Kageyama R. Mash1 and Math3 are required for development of branchiomotor neurons and maintenance of neural progenitors. J Neurosci. 2005;25:5857–5865. doi: 10.1523/JNEUROSCI.4621-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil V, Hirsch MR, Jouve C, Brunet JF, Goridis C. The role of Phox2b in synchronizing pan-neuronal and type-specific aspects of neurogenesis. Development. 2002;129:5241–5253. doi: 10.1242/dev.129.22.5241. [DOI] [PubMed] [Google Scholar]
- Pattyn A, Simplicio N, van Doorninck JH, Goridis C, Guillemot F, Brunet JF. Mash1/Ascl1 is required for the development of central serotonergic neurons. Nat Neurosci. 2004;7:589–595. doi: 10.1038/nn1247. [DOI] [PubMed] [Google Scholar]
- Dubreuil V, Hirsch MR, Pattyn A, Brunet JF, Goridis C. The Phox2b transcription factor coordinately regulates neuronal cell cycle exit and identity. Development. 2000;127:5191–5201. doi: 10.1242/dev.127.23.5191. [DOI] [PubMed] [Google Scholar]
- Brunet JF, Goridis C. Phox2b and the homeostatic brain. In: Gaultier C, editor. Genetic basis for respiratory control diseases. New York , Springer; 2008. pp. 25–38. [Google Scholar]
- Grillet N, Dubreuil V, Dufour HD, Brunet JF. Dynamic expression of RGS4 in the developing nervous system and regulation by the neural type-specific transcription factor Phox2b. J Neurosci. 2003;23:10613–10621. doi: 10.1523/JNEUROSCI.23-33-10613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caroni P. Intrinsic neuronal determinants that promote axonal sprouting and elongation. Bioessays. 1997;19:767–775. doi: 10.1002/bies.950190906. [DOI] [PubMed] [Google Scholar]
- Benowitz LI, Routtenberg A. GAP-43: an intrinsic determinant of neuronal development and plasticity. Trends Neurosci. 1997;20:84–91. doi: 10.1016/s0166-2236(96)10072-2. [DOI] [PubMed] [Google Scholar]
- Schneider MR, Wolf E, Hoeflich A, Lahm H. IGF-binding protein-5: flexible player in the IGF system and effector on its own. J Endocrinol. 2002;172:423–440. doi: 10.1677/joe.0.1720423. [DOI] [PubMed] [Google Scholar]
- Gurok U, Steinhoff C, Lipkowitz B, Ropers HH, Scharff C, Nuber UA. Gene expression changes in the course of neural progenitor cell differentiation. J Neurosci. 2004;24:5982–6002. doi: 10.1523/JNEUROSCI.0809-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng SR, Lanier J, Fedtsova N, Turner EE. Coordinated regulation of gene expression by Brn3a in developing sensory ganglia. Development. 2004;131:3859–3870. doi: 10.1242/dev.01260. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Hatakeyama J, Ohsawa R. Roles of bHLH genes in neural stem cell differentiation. Exp Cell Res. 2005;306:343–348. doi: 10.1016/j.yexcr.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Kobayashi T. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development. 2007;134:1243–1251. doi: 10.1242/dev.000786. [DOI] [PubMed] [Google Scholar]
- Korinek V, Barker N, Willert K, Molenaar M, Roose J, Wagenaar G, Markman M, Lamers W, Destree O, Clevers H. Two members of the Tcf family implicated in Wnt/beta-catenin signaling during embryogenesis in the mouse. Mol Cell Biol. 1998;18:1248–1256. doi: 10.1128/mcb.18.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arce L, Yokoyama NN, Waterman ML. Diversity of LEF/TCF action in development and disease. Oncogene. 2006;25:7492–7504. doi: 10.1038/sj.onc.1210056. [DOI] [PubMed] [Google Scholar]
- Kawano Y, Kypta R. Secreted antagonists of the Wnt signalling pathway. J Cell Sci. 2003;116:2627–2634. doi: 10.1242/jcs.00623. [DOI] [PubMed] [Google Scholar]
- Rodriguez J, Esteve P, Weinl C, Ruiz JM, Fermin Y, Trousse F, Dwivedy A, Holt C, Bovolenta P. SFRP1 regulates the growth of retinal ganglion cell axons through the Fz2 receptor. Nat Neurosci. 2005;8:1301–1309. doi: 10.1038/nn1547. [DOI] [PubMed] [Google Scholar]
- Esteve P, Bovolenta P. Secreted inducers in vertebrate eye development: more functions for old morphogens. Curr Opin Neurobiol. 2006;16:13–19. doi: 10.1016/j.conb.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Schepers GE, Teasdale RD, Koopman P. Twenty pairs of sox: extent, homology, and nomenclature of the mouse and human sox transcription factor gene families. Dev Cell. 2002;3:167–170. doi: 10.1016/s1534-5807(02)00223-x. [DOI] [PubMed] [Google Scholar]
- Kasimiotis H, Myers MA, Argentaro A, Mertin S, Fida S, Ferraro T, Olsson J, Rowley MJ, Harley VR. Sex-determining region Y-related protein SOX13 is a diabetes autoantigen expressed in pancreatic islets. Diabetes. 2000;49:555–561. doi: 10.2337/diabetes.49.4.555. [DOI] [PubMed] [Google Scholar]
- Melichar HJ, Narayan K, Der SD, Hiraoka Y, Gardiol N, Jeannet G, Held W, Chambers CA, Kang J. Regulation of gammadelta versus alphabeta T lymphocyte differentiation by the transcription factor SOX13. Science. 2007;315:230–233. doi: 10.1126/science.1135344. [DOI] [PubMed] [Google Scholar]
- Nguyen L, Besson A, Roberts JM, Guillemot F. Coupling cell cycle exit, neuronal differentiation and migration in cortical neurogenesis. Cell Cycle. 2006;5:2314–2318. doi: 10.4161/cc.5.20.3381. [DOI] [PubMed] [Google Scholar]
- Stoeckli ET, Landmesser LT. Axonin-1, Nr-CAM, and Ng-CAM play different roles in the in vivo guidance of chick commissural neurons. Neuron. 1995;14:1165–1179. doi: 10.1016/0896-6273(95)90264-3. [DOI] [PubMed] [Google Scholar]
- Gross MK, Dottori M, Goulding M. Lbx1 specifies somatosensory association interneurons in the dorsal spinal cord. Neuron. 2002;34:535–549. doi: 10.1016/s0896-6273(02)00690-6. [DOI] [PubMed] [Google Scholar]
- Muller T, Brohmann H, Pierani A, Heppenstall PA, Lewin GR, Jessell TM, Birchmeier C. The homeodomain factor lbx1 distinguishes two major programs of neuronal differentiation in the dorsal spinal cord. Neuron. 2002;34:551–562. doi: 10.1016/s0896-6273(02)00689-x. [DOI] [PubMed] [Google Scholar]
- Melkonyan HS, Chang WC, Shapiro JP, Mahadevappa M, Fitzpatrick PA, Kiefer MC, Tomei LD, Umansky SR. SARPs: a family of secreted apoptosis-related proteins. Proc Natl Acad Sci U S A. 1997;94:13636–13641. doi: 10.1073/pnas.94.25.13636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellies DL, Church V, Francis-West P, Lumsden A. The WNT antagonist cSFRP2 modulates programmed cell death in the developing hindbrain. Development. 2000;127:5285–5295. doi: 10.1242/dev.127.24.5285. [DOI] [PubMed] [Google Scholar]
- Homma S, Yaginuma H, Oppenheim RW. Programmed cell death during the earliest stages of spinal cord development in the chick embryo: a possible means of early phenotypic selection. J Comp Neurol. 1994;345:377–395. doi: 10.1002/cne.903450305. [DOI] [PubMed] [Google Scholar]
- Pevny L, Placzek M. SOX genes and neural progenitor identity. Curr Opin Neurobiol. 2005;15:7–13. doi: 10.1016/j.conb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Skene JH. Axonal growth-associated proteins. Annu Rev Neurosci. 1989;12:127–156. doi: 10.1146/annurev.ne.12.030189.001015. [DOI] [PubMed] [Google Scholar]
- Aigner L, Arber S, Kapfhammer JP, Laux T, Schneider C, Botteri F, Brenner HR, Caroni P. Overexpression of the neural growth-associated protein GAP-43 induces nerve sprouting in the adult nervous system of transgenic mice. Cell. 1995;83:269–278. doi: 10.1016/0092-8674(95)90168-x. [DOI] [PubMed] [Google Scholar]
- Strittmatter SM, Fankhauser C, Huang PL, Mashimo H, Fishman MC. Neuronal pathfinding is abnormal in mice lacking the neuronal growth cone protein GAP-43. Cell. 1995;80:445–452. doi: 10.1016/0092-8674(95)90495-6. [DOI] [PubMed] [Google Scholar]
- Donovan SL, Mamounas LA, Andrews AM, Blue ME, McCasland JS. GAP-43 is critical for normal development of the serotonergic innervation in forebrain. J Neurosci. 2002;22:3543–3552. doi: 10.1523/JNEUROSCI.22-09-03543.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Mani S, Donovan SL, Schwob JE, Meiri KF. Growth-associated protein-43 is required for commissural axon guidance in the developing vertebrate nervous system. J Neurosci. 2002;22:239–247. doi: 10.1523/JNEUROSCI.22-01-00239.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey D, Laux T, Xu L, Schneider C, Caroni P. Shared and unique roles of CAP23 and GAP43 in actin regulation, neurite outgrowth, and anatomical plasticity. J Cell Biol. 2000;149:1443–1454. doi: 10.1083/jcb.149.7.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill BJ, Pasolli HA, Polak L, Rendl M, Garcia-Garcia MJ, Anderson KV, Fuchs E. Tcf3: a transcriptional regulator of axis induction in the early embryo. Development. 2004;131:263–274. doi: 10.1242/dev.00935. [DOI] [PubMed] [Google Scholar]
- Pattyn A, Goridis C, Brunet JF. Specification of the central noradrenergic phenotype by the homeobox gene Phox2b. Mol Cell Neurosci. 2000;15:235–243. doi: 10.1006/mcne.1999.0826. [DOI] [PubMed] [Google Scholar]
- Terry K, Magan H, Baranski M, Burrus LW. Sfrp-1 and sfrp-2 are expressed in overlapping and distinct domains during chick development. Mech Dev. 2000;97:177–182. doi: 10.1016/s0925-4773(00)00407-x. [DOI] [PubMed] [Google Scholar]
- Muroyama Y, Fujihara M, Ikeya M, Kondoh H, Takada S. Wnt signaling plays an essential role in neuronal specification of the dorsal spinal cord. Genes Dev. 2002;16:548–553. doi: 10.1101/gad.937102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve P, Trousse F, Rodriguez J, Bovolenta P. SFRP1 modulates retina cell differentiation through a beta-catenin-independent mechanism. J Cell Sci. 2003;116:2471–2481. doi: 10.1242/jcs.00452. [DOI] [PubMed] [Google Scholar]
- Bylund M, Andersson E, Novitch BG, Muhr J. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat Neurosci. 2003;6:1162–1168. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- Sandberg M, Kallstrom M, Muhr J. Sox21 promotes the progression of vertebrate neurogenesis. Nat Neurosci. 2005;8:995–1001. doi: 10.1038/nn1493. [DOI] [PubMed] [Google Scholar]
- Bergsland M, Werme M, Malewicz M, Perlmann T, Muhr J. The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes Dev. 2006;20:3475–3486. doi: 10.1101/gad.403406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Bagheri-Fam S, Harley VR. SOX13 is up-regulated in the developing mouse neuroepithelium and identifies a sub-population of differentiating neurons. Brain Res Dev Brain Res. 2005;157:201–208. doi: 10.1016/j.devbrainres.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Pattyn A, Morin X, Cremer H, Goridis C, Brunet JF. The homeobox gene Phox2b is essential for the development of autonomic neural crest derivatives. Nature. 1999;399:366–370. doi: 10.1038/20700. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Lo LC, Johnson JE, Auerbach A, Anderson DJ, Joyner AL. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75:463–476. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- Tanaka TS, Jaradat SA, Lim MK, Kargul GJ, Wang X, Grahovac MJ, Pantano S, Sano Y, Piao Y, Nagaraja R, Doi H, Wood WH, 3rd, Becker KG, Ko MS. Genome-wide expression profiling of mid-gestation placenta and embryo using a 15,000 mouse developmental cDNA microarray. Proc Natl Acad Sci U S A. 2000;97:9127–9132. doi: 10.1073/pnas.97.16.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Herrero J, Al-Shahrour F, Diaz-Uriarte R, Mateos A, Vaquerizas JM, Santoyo J, Dopazo J. GEPAS: A web-based resource for microarray gene expression data analysis. Nucleic Acids Res. 2003;31:3461–3467. doi: 10.1093/nar/gkg591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida T, Ensini M, Morton SB, Baldassare M, Edlund T, Jessell TM, Pfaff SL. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79:957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes down-regulated in Phox2bLacZ/LacZ embryos. A list of the 51 genes that were found to be significantly down-regulated in the ventral r4 of Phox2bLacZ/LacZ embryos according to the criteria presented in Materials and methods (SAM score above 3.5). The expression of the genes written in red was tested by ISH in Phox2bLacZ/+ and Phox2bLacZ/LacZembryos.
Genes up-regulated in Phox2bLacZ/LacZ embryos. A list of the 23 genes that were found to be significantly up-regulated in the ventral r4 of Phox2bLacZ/LacZ embryos according to the criteria presented in Materials and methods (SAM score below -3.5). The expression of the genes written in red was tested by ISH in Phox2bLacZ/+ and Phox2bLacZ/LacZembryos.