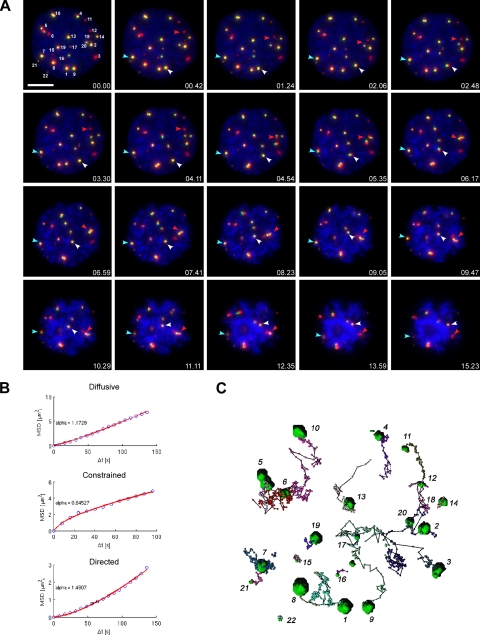
Figure 2.
4D tracking of PML NBs from prophase to prometaphase in living cells. (A) Double stable cells, CpYs68, stably expressing PML-ECFP and EYFP-Sp100′. Chromatin was visualized by Hoechst 33342 staining. A 7-μm Z-stack of 0.5-μm steps were collected in the CFP and YFP channels every 8 s for 26 min. One Z-section at the middle of the cell was collected in the Hoechst channel, at each time point, as the reference image for DNA. The 3D-projected still images of selected time points are shown here. Each PML NB was manually assigned a number and tracked by TIKAL 4D viewer to determine their displacement. The examples of PML NBs with different motions are indicated by colored arrowheads. The red arrowhead, directed motion. White arrowhead, diffusive motion. Blue arrowhead, constrained motion. Red, pseudocolored PML-ECFP. Green, EYFP-Sp100′. Blue, DNA stained by Hoechst 33342. Bar, 10 μm. (B) The trajectories of bodies were exported into MATLAB to calculate the MSD, which is plotted in the y-axis against Δt in the x-axis. The MSD is fitted to the power law MSD = 6DΔtα, with α as the coefficient of anomalous diffusion. A nonlinear relationship between the MSD and Δt indicates nonstandard diffusion. Classification into movement types is performed according to α ∼ 1.5 for directed movement, α ∼ 1 for simple diffusion, and α ∼ 0.5 for constrained diffusion. The representative MSD plot of each movement type is displayed here with the actual data (blue circles) and the fitted curve (red line). (C) Overview of the trajectory of each PML NB in this 4D image data set is summarized by ball-and-stick representations to show the displacement of each body. The initial position of each PML NB is indicated by a green volume.