Abstract
DNA methylation at cytosines is a widely studied epigenetic modification. Methylation is commonly detected using bisulfite modification of DNA followed by PCR and additional techniques such as restriction digestion or sequencing. These additional techniques are either laborious, require specialized equipment, or are not quantitative. Here we describe a simple algorithm that yields quantitative results from analysis of conventional four-dye-trace sequencing. We call this method Mquant and we compare it with the established laboratory method of combined bisulfite restriction assay (COBRA). This analysis of sequencing electropherograms provides a simple, easily applied method to quantify DNA methylation at specific CpG sites.
INTRODUCTION
Methylation of cytosines in DNA is an epigenetic modification in vertebrates, higher plants and some other eukaryotes. It is strongly associated with gene silencing, and its gene- and site-specific quantification is important to understand epigenetic changes in biology including in development, behavior, cancer and aging (1–8).
Site-specific DNA methylation can be quantified by numerous methods, most of which use restriction digestion and/or bisulfite treatment (9–23). Some of these methods are limited to just one or a few sites. Several methods use genomic sequencing to quantify methylation over stretches of DNA up to a few hundred nucleotides. Each of these require specialized techniques or equipment that are not widely used or widely available (10,13,16,18,19). Bisulfite genomic sequencing (BGS) and related bisulfite-based techniques (9,24) are some of the most useful methods to detect DNA methylation. Capillary electrophoresis methods producing four-dye-trace electropherograms are widely used to detect methylation with BGS. However, this method is not quantitative without subcloning, sequencing and averaging each sample (25–27) or without use of complex, specialized algorithms (16). Recently, Dikow et al. (12) described a simple way to quantify DNA methylation from BGS four-dye-trace electropherograms, but they show the maximum mean signal generated to be just over 80% methylation, and they suggest that quantification by bisulfite treatment may be intrinsically problematic. They presented data showing that a specialized, nonbisulfite technique (12,17) was more accurate.
Here we report a BGS analysis method that quantifies methylation at any particular site by subtracting the thymine signal at that site from the average signal of 10 surrounding thymine peaks. Results with this method are highly correlated with, and give similar values as, DNA methylation levels measured on the same sites with the well-established COBRA assay. Both this new method and COBRA measure levels of methylation in the midrange and near the extremes of 0 and 100%. These results show that DNA methylation can be quantified by simple analysis of BGS four-dye-trace electropherograms.
MATERIALS AND METHODS
DNA extraction and in vitro methylation
DNA was extracted from mouse tissues using an Epicentre MasterPure DNA purification kit (Epicentre Biotechnologies, Madison, WI, USA) according to the manufacturer's recommendations with minor modifications. We added a phenol (Amresco, Solon OH, USA) extraction step and a 1-bromo-3-chloropropane (Molecular Research Center, Inc. Cincinnati, OH, USA) extraction step just prior to isopropanol precipitation. Purified DNA was washed with Tris–EDTA buffer in Montage centrifugal filters (Millipore, Bedford, MA, USA). In some cases DNA was methylated in vitro with SssI (CpG) methylase according to the manufacturer's instructions (New England Biolabs Inc, Ipswich, MA, USA) except that DNA was washed in a centrifugal filter and reacted a second time (11) to assure complete methylation.
Bisulfite modification of DNA
DNA was sodium-bisulfite modified with an Epitect Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. For each bisulfite modification we used 300 ng of DNA. We stored bisulfite-treated DNA at −20°C.
PCR
PCR was performed using a Hot Star Taq DNA polymerase kit (Qiagen, Valencia, CA, USA). Each 25-μl PCR reaction included 0.65 U of Hot Star Taq polymerase, 0.22 mM Promega dNTP mix (Promega, Madison, WI, USA) and 0.8 μM of each primer. The sequences amplified were from the mouse Avy allele of agouti (28) (Genbank AR302985). Bisulfite-modified genomic DNA was amplified by nested PCR using two sets of primers for the Avy allele similar to that described by Rakyan et al. (29).
The first PCR reaction was carried out with 10 ng of DNA using the amplification profile: 1 cycle at 80°C for 1 min, 1 cycle at 94°C for 1 min; 2 cycles at (95°C for 1 min, 64°C for 1 min, 72°C for 1 min); 2 cycles at (95°C for 1 min, 63°C for 1 min, 72°C for 1 min); 2 cycles at (95°C for 1 min, 62°C for 1 min, 72°C for 1 min); 2 cycles at (95°C for 1 min, 61°C for 1 min, 72°C for 1 min); 40 cycles at (95°C for 1 min, 60°C for 1 min, 72°C for 1 min); 72°C for 5 min and cooling to 4°C.
The forward primer 5′-TGCGATAAAGTTTTATTTTTAT-3′ and reverse primer 5′-GTTGTGTTTCGTTTTGTTTTTTTTTT-3′ used for the first reaction were designed using MethPrimer web software (30) (http://www.urogene.org/methprimer/). A second, nested, PCR was then performed on 1 μl of the amplificate using the upstream and downstream Avy primers (372-bp PCR product) or the upstream and internal Avy primers (307-bp PCR product) of Rakyan et al. (29) with the following cycling conditions: 1 cycle at 80°C for 1 min, 1 cycle at 94°C for 1 min; 2 cycles at (95°C for 1 min, 63°C for 1 min, 72°C for 1 min); 2 cycles at (95°C for 1 min, 62°C for 1 min, 72°C for 1 min); 2 cycles at (95°C for 1 min, 61°C for 1 min, 72°C for 1 min); 2 cycles at (95°C for 1 min, 60°C for 1 min, 72°C for 1 min); 40 cycles at (95°C for 1 min, 59°C for 1 min, 72°C for 1 min); 72°C for 5 min and cooling to 4°C. The PCR products were electrophoresed through a 2% agarose gel, stained with ethidium bromide and digitally imaged under UV light using a transilluminator, video camera and LabWorks image acquisition and analysis software (Ultra-Violet Products, Upland CA, USA).
BGS
To eliminate primers and dNTPs, we treated PCR products with exonuclease I (Epicentre, Madison WI, USA) and shrimp alkaline phosphatase (Roche, Nutley, NJ, USA) (31) at 37°C for 60 min followed by 85°C for 15 min. We then concentrated and washed these using Montage centrifugal filters (Millipore, Bedford, MA, USA) according to the manufacturer's recommendations. The PCR products were sequenced using the nested upstream primer (Avy forward primer) (29) at the UAMS DNA Sequencing Core Facility using a Model 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) and a Big Dye terminator sequencing kit.
Combined bisulfite restriction assay (COBRA)
For COBRA analysis (22,23) PCR products were digested with 20 U of restriction enzyme TaqalphaI (TCGA), HpyCH4IV (ACGT) or AciI (GGCG)(New England Biolabs, Ipswich, MA, USA). Each of these enzymes has just one site in the bisulfite-converted sequence when the original genomic sequence was methylated, and no site in the bisulfite-converted sequence when the original genomic sequence was unmethylated. For digestion, a 10- to 20-fold excess of enzyme was used for 2 h, but digestion was otherwise according to the manufacturer's instructions. The digested PCR products were separated by gel electrophoresis using 3% GenePure high-resolution agarose (ISC BioExpress, Kaysville, UT, USA) and stained with ethidium bromide. Gels were imaged as described earlier and the images saved as TIFF files.
For COBRA electrophoresis the amount of digest analyzed was kept low so that the bands were in a gray level (in an approximately linear range) but high enough that they gave a substantial signal. The undigested band and the largest-size digested band were used to quantify methylation because the smaller-size digest bands sometimes did not give a substantial signal. Even at extremes of methylation (near 0 or 100%), at least one band, the undigested or the largest digested band, gave a substantial signal.
Digital images were scanned with Scion Image software (Scion Corporation, Frederick, MD, USA, http://www.scioncorp.com/pages/scion_image_windows.htm) to measure density. Density ratios of a major digested band to the undigested band were used to calculate the relative copy numbers of fragments and subsequently the percent methylation (11,23).
Peak area determination from sequencing electropherograms
The ab1 files from sequencing were processed using Phred (32,33) (http://www.phrap.org/) or BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Sequences were not used if they had substantial artifacts e.g. if more than one T (that had not been a C of a CpG prior to bisulfite) in the region used to quantify methylation showed more than 10% C. The Phred output or BioEdit trace value output were used to read and quantify primary and secondary peaks in the electropherograms. Calculations were performed in Excel (Microsoft, Redmond, WA, USA). Peak areas were determined by summing peak trace values. Phred automatically sums the trace values of peaks from baseline (32) and we did the same with trace values from Bioedit after pasting them into Excel spreadsheets. Virtually all baselines between peaks had one or more trace values of zero which allows peak area determination by simple summing of peak trace values.
Algorithm for quantification of DNA methylation
DNA methylation levels were quantified from sequencing electropherogram trace values using the following algorithm that we call Mquant.
First, the target CpG site was chosen.
Second, the mean T area (T bar) from 10 Ts surrounding the target CpG site was determined. Ts used to calculate T bar were at least 10 times the area of their secondary base (C, G or A). In our electropherograms, secondary bases were mainly sequencing noise. Thus the Ts used to calculate T bar had a T signal-to-noise ratio of 10 or better with respect to their secondary peak. An equal number of Ts from each side of the target CpG (i.e. 5 on each side) were used.
Third, the area of the T at the target CpG site was subtracted from T bar to yield delta T (i.e. T bar − CpG T = delta T).
Fourth, the level of methylation on the site was calculated as the proportion, (delta T)/(T bar), or the percentage, 100 X (delta T)/(T bar).
Data analysis
Percents methylation by the Mquant and COBRA methods were compared via regression plots using Origin (OriginLab, Northhampton, MA, USA) and via Bland-Altman plots using Excel (Microsoft, Redmond, WA, USA). Regression was used to calculate slopes, intercepts and coefficients of correlation between methods, whereas Bland-Altman plots (34,35) were used to determine means and standard deviations (SDs) of the difference in percents methylation by each method. Bland-Altman 95% Limits of Agreement (95% LoAs) were calculated as the mean ± 2 SD of the difference in percents methylation by each method, and indicate the limits between which ∼95% of the difference in percents methylation would be expected to fall under a ‘normal’ distribution. In some cases we did multiple analyses of single amplificates for which we determined SD, the root mean square error (RMSE) and the coefficient of variation (CV) as measures of run-to-run variation within each experiment.
RESULTS
We have developed a method to quantify DNA methylation from BGS electropherograms. This method uses and extends previously published methods of sequence analysis (12,32,33,36) so that we can readily quantify the methylation at a particular site using the data from four-dye-trace electropherograms from fluorescent dye terminator sequencing. This allows us to quantify the percent methylation of numerous CpG sites in an electrophoretogram and to validate these levels at specific sites using COBRA assays (23). This method can greatly speed determinations of DNA methylation.
COBRA distinguishes between C and T bases using sequence-specific restriction enzymes as measured by the intensities of bands on a gel after DNA fragments are separated by electrophoresis (22,23). BGS, as used here, distinguishes between C and T bases by different fluorescent dyes on each base at specific positions after separation by capillary electrophoresis. At a target CpG site, T is measured directly and C is measured indirectly as the mean intensity of surrounding Ts minus the intensity of T at the target site (which is shared by C and T).
We used bisulfite PCR amplificates from 45 independent DNA samples containing sites with DNA methylation levels that varied between 0 and 100%. With these, we compared the bisulfite-based techniques of COBRA with a quantitative version of BGS that we call Mquant. The agouti allele region we used contains nine CpGs that can be sequenced reliably with the primer sets used. Three of these CpGs are in restriction sites, and were analyzed by both COBRA and Mquant.
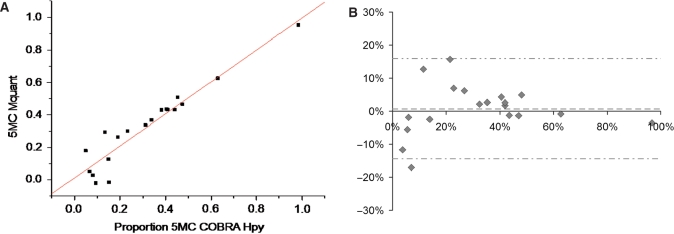
A total of 61 COBRA and 61 Mquant determinations were made (from 45 PCR amplificates) to test for agreement between COBRA and Mquant. Each COBRA and corresponding Mquant was performed on the same bisulfite PCR amplificate. Figure 1 shows a COBRA gel for measuring methylation of an HpyCH4IV site (ACGT) and the corresponding site in the sequencing electropherogram. In this and other COBRA gels the amounts of digest loaded were in a moderate range so that the bands were at a gray level (in an approximately linear range) and still gave a substantial signal. Figure 2A is a regression plot of COBRA values versus electropherogram values at the ACGT site, and shows a high correlation (0.95) between values measured by the two methods. Figure 2B is a Bland-Altman plot for this same data. The mean (SD) difference between COBRA and Mquant values for the HpyCH4IV site was +0.72% (7.6%), indicating little evidence for bias (P = 0.68) between methods. The outside horizontal lines of Figure 2B show the Bland-Altman 95%LoA's, which are (−14.4%, +15.9%) for the ACGT site. The mean values of percent methylation for COBRA and Mquant were 31.6 and 32.4%, respectively. Overall, these results show that the two methods tend to agree well at the HpyCH4IV site.
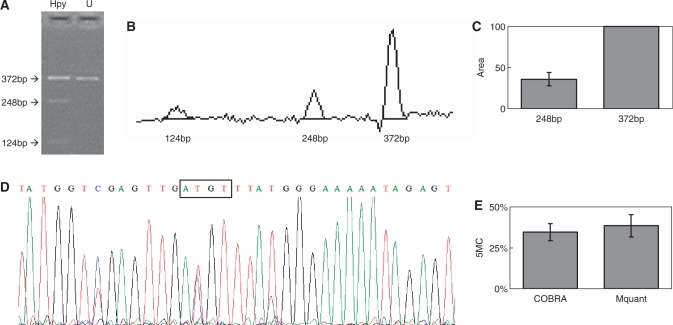
Figure 1.
Gel electrophoresis for quantification of DNA methylation by COBRA, a scan of this gel and a corresponding electropherogram from the same amplificate analyzed by Mquant. (A) A bisulfite PCR product was digested with HpyCH4IV (ACGT) to give three bands of 372, 248 and 124 bp. The 372-bp band is undigested DNA (representing unmethylated DNA that now has an ATGT site). The smaller bands represent methylated DNA whose single HpyCH4IV site was cleaved. The digested DNA lane is marked Hpy and the undigested DNA lane is marked U. (B) A scan of the digested lane of this gel. (C) The peak areas of the 372- and 248-bp bands from four determinations were quantified and their relative peak areas are shown (with 372 bp areas normalized to 100). (D) The T trace of the electropherogram was analyzed by Mquant as described in the text. (E) The relative copy numbers of the 372- and 248-bp bands were used to calculate the percent DNA methylation (5MC) of the original DNA. The percent methylation by Mquant is also shown. Analyses of this individual PCR amplificate gave a mean percent methylation (±SD) by COBRA of 34.6 ± 5.2% with a CV of 15% (n = 4) and a mean percent methylation by Mquant of 38.4 ± 6.8 with a CV of 18% (n = 4). The differences in the two methods for this amplificate are not statistically significant (P = 0.40) and are <4% methylation (35 versus 38%).
Figure 2.
(A) A plot of the percent DNA methylation on the CpG of an ACGT site from 19 different DNA samples determined by COBRA versus the percent methylation on this same site determined by peak areas on forward sequence electropherograms. The results of the two methods were highly correlated (R = 0.95, P < 10−9) and in good agreement (estimate ± standard error = 0.98 ± 0.079 for slope, and 0.012 ± 0.031 for y-intercept). The average percent methylation measured by COBRA and Mquant were both 32%. (B) A Bland-Altman plot of the data shown in (A). The vertical axis shows the difference in methylation values measured by the two methods (Mquant minus COBRA), whereas the horizontal axis shows the average methylation value measured by the two. The mean (SD) of the difference between methods was +0.72% (7.6%), indicating little evidence for bias between methods (P = 0.68). The center dashed horizontal line shows the mean difference, while the outside horizontal lines show the 95%LoAs (at −14.4%, +15.9%).
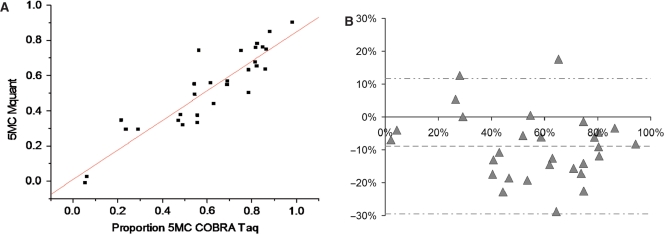
Figures 3 through 4B show analogous results for the TaqalphaI site (TCGA). The correlation between methylation levels measured by the two methods was somewhat lower (0.91). The mean (SD) difference between values measured by COBRA versus Mquant was −8.9% (10.3%), indicating statistically significant evidence (P < 0.0001) of a bias toward lower values as measured by Mquant compared to COBRA. The 95%LoAs were (−29.5%, +11.7%) for the TCGA site. The mean values of percent methylation for COBRA and Mquant were 61% and 52%, respectively. Although the bias was statistically significant at the TaqalphaI site, it was nevertheless under 10% methylation.
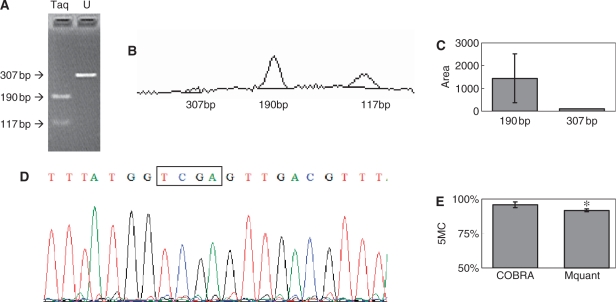
Figure 3.
Gel electrophoresis for quantification of DNA methylation by COBRA, a scan of this gel and a corresponding electropherogram from the same amplificate analyzed by Mquant. (A) A bisulfite PCR product was digested with TaqalphaI (TCGA) to give three bands of 307, 190 and 117 bp. The 307-bp band is undigested DNA (representing unmethylated DNA that now has a TTGA site). The smaller bands represent methylated DNA whose single TaqalphaI site was cleaved. The peak areas of the 307- and 190-bp bands were quantified and their relative copy numbers were calculated and used to calculate the percent methylation of the original DNA. The digested DNA lane is marked Taq and the undigested DNA lane is marked U. (B) A scan of the digested lane of this gel. (C) The peak areas of the 307- and 190-bp bands from five determinations were quantified and their relative peak areas are shown (with 307-bp areas normalized to 100). (D) The T trace of the electropherogram was analyzed by Mquant as described in the text. (E) The relative copy numbers of the 307- and 190-bp bands were used to calculate the percent DNA methylation of the original DNA. The percent methylation by Mquant is also shown. Analyses of this individual PCR amplificate gave a mean percent methylation (±SD) by COBRA of 95.7 ± 2.1 with a CV of 2.2% (n = 5) and a mean percent methylation by Mquant of 91.7 ± 1.0 with a CV of 1.1% (n = 5). The differences in the two methods for this amplificate are statistically significant [P < 0.01, marked with an asterisk in (E)] but differ by only 4% methylation (96 versus 92%). The standard deviation (shown as error bars) is large in (C) because it includes the variation (2.1%) in the percent of unmethylated site (4.3%).
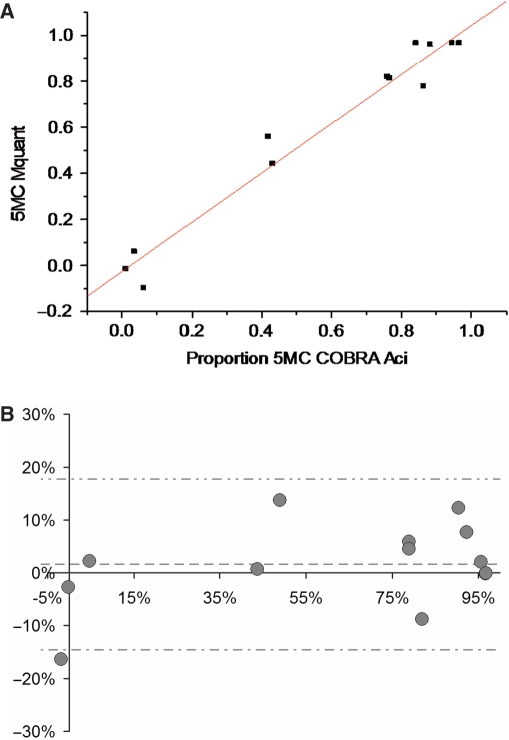
Figures 5 through 6B show analogous results for the AciI site (GCGG). The correlation between methylation levels measured by the two methods was high (0.98). The mean (SD) difference between values measured by COBRA versus Mquant was 1.6% (8.1%), indicating little evidence for bias (P = 0.48) between methods. The 95%LoAs were (−14.6%, +17.8%) for the GCGG site. The mean values of percent methylation for COBRA and Mquant were 61% and 63%, respectively. Overall, these results show that the two methods tend to agree well at the AciI site.
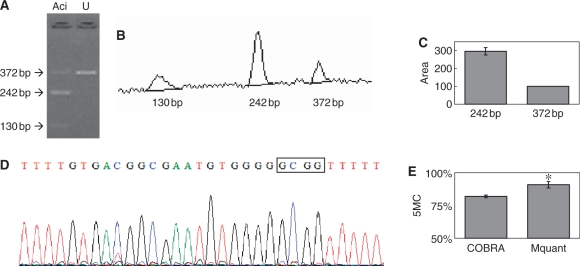
Figure 5.
Gel electrophoresis for quantification of DNA methylation by COBRA, a scan of this gel and a corresponding electropherogram from the same amplificate analyzed by Mquant. (A) A bisulfite PCR product was digested with AciI (GCGG) to give three bands of 372, 242 and 130 bp. The 372-bp band is undigested DNA (representing unmethylated DNA that now has a GTGG site). The smaller bands represent methylated DNA whose single AciI site was cleaved. The peak areas of the 372- and 242-bp bands were quantified and their relative copy numbers were calculated and used to calculate the percent methylation of the original DNA. The digested DNA lane is marked Aci and the undigested DNA lane is marked U. (B) A scan of the digested lane of this gel. (C) The peak areas of the 372- and 242-bp bands from four determinations were quantified and their relative peak areas are shown (with 372-bp areas normalized to 100). (D) The T trace of the electropherogram was analyzed by Mquant as described in the text. (E) The relative copy numbers of the 372- and 242-bp bands were used to calculate the percent DNA methylation of the original DNA. The percent methylation by Mquant is also shown. Analyses of this individual PCR amplificate gave a mean percent methylation (± SD) by COBRA of 82.0 ± 1.0 with a CV of 1.2% (n = 4) and a mean percent methylation by Mquant of 90.9 ± 2.5 with a CV of 2.7% (n = 4). The differences in the two methods for this amplificate are statistically significant [P < 0.003, marked with an asterisk in (E)] but only differ by 9% methylation (82 versus 91%).
We made estimates of C to T conversion levels and general noise levels in the electropherograms. First we measured the C level under Ts from non-CpG Cs. These levels were small and indicated a conversion rate of >93–97%. Next we measured the levels of other bases (G and A) under Ts from non-CpG Cs. Levels of G and A were similar to those of C indicating that a substantial amount of C level may come from sequencing noise and not from incomplete C to T conversion. In any case, C to T conversion levels appear to be between 93 and 100%.
We tested the number of Ts used to calculate T bar on the calculated DNA methylation level (data not shown) and on correlations with COBRA (Table 1). We tested the use of 2 Ts (one on each side), 4 Ts (two on each side) and so on, up to 20 Ts (10 on each side). In all cases R was >0.90 and all correlations were highly significant (10−12 < P < 10−7). For AciI and TaqalphaI sites the number of Ts between 2 and 20 had little effect on R (0.97–0.98 and 0.90–0.92, respectively). For HpyCH4IV sites R was 0.91 using 2 or 4 Ts and 0.93–0.95 using 8–20 Ts.
Table 1.
The effects of T numbers on correlations and P-values when Mquant is compared with COBRA
| 2T | 4T | 6T | 8 to 20T | |
|---|---|---|---|---|
| HpyCH4IV | ||||
| R | 0.91 | 0.91 | 0.92 | 0.93–0.95 |
| P | 2.6 × 10−8 | 2.9 × 10−8 | 1.0 × 10−8 | <2 × 10−9 |
| Taqalpha1 | ||||
| R | 0.90 | 0.91 | 0.92 | 0.91–0.92 |
| P | 1.6 × 10−11 | 1.0 × 10−11 | 2.2 × 10−12 | <8 × 10−12 |
| AciI | ||||
| R | 0.97 | 0.97 | 0.97 | 0.98 |
| P | 2.2 × 10−8 | 5.2 × 10−8 | 1.6 × 10−8 | <6 × 10−9 |
The data shown in Figures 2A, 4A and 6A are a large collection of single COBRA and Mquant determinations from a large number of amplificates. Additionally, to assess run-to-run reproducibility, 10 amplificates were subsampled three to five times and assayed by both methods. The resulting data were analyzed statistically via one-way ANOVA on the parent amplificates in order to obtain the ANOVA RMSE, which estimates the common standard deviation (SD) of the amplificate replications about their respective mean values. For COBRA, RMSEs were 4.3, 1.5 and 1.0% for Hpy, Taq and Aci, respectively. For Mquant, RMSEs were 4.5, 1.8 and 1.6% for Hpy, Taq and Aci, respectively. For the three sites combined, COBRA RMSE was 2.7% and Mquant RMSE was 3.0%. These estimates of SD are low for most methylation levels. For example, an SD of 3.0% is reasonable for a measured methylation level of 90, 50 or even 20%. Only when methylation levels were very low (e.g. <10%) was the SD a substantial proportion of the measured methylation level. Overall, SDs were low, indicating that each method is highly reproducible.
Figure 4.
(A) A plot of the percent DNA methylation on the CpG of a TCGA site from 29 different DNA samples determined by COBRA versus the percent methylation on this same site determined by peak areas on forward sequence electropherograms. The results of the two methods are highly correlated (R = 0.91, P = 7.0 × 10−12), but in rather poor agreement (estimate ± standard error = 0.84 ± 0.073 for the slope, and 0.009 ± 0.048 for the y-intercept). The average percent methylation measured by COBRA and Mquant were 61% and 52%, respectively. (B) A Bland-Altman plot of the data shown in (A). The vertical axis shows the difference in methylation values measured by the two methods (Mquant minus COBRA), whereas the horizontal axis shows the average methylation value measured by the two. The mean (SD) of the difference between methods was −8.9% (10.3%), indicating statistically significant evidence (P < 0.0001) of a bias toward lower values as measured by Mquant compared to COBRA. The center dashed horizontal line shows the mean difference, while the outside horizontal lines show the 95%LoAs (at −29.5%, +11.7%).
Figure 6.
(A) A plot of the percent DNA methylation on the CpG of a GCGG site from 13 different DNA samples determined by COBRA versus the percent methylation on this same site determined by peak areas on forward sequence electropherograms. The results of the two methods are highly correlated (R = 0.98, P = 2.7 × 10−9) and in reasonably close agreement (estimate ± standard error = 1.07 ± 0.062 for the slope, and −0.026 ± 0.044 for the y-intercept). The average percent methylation measured by COBRA and Mquant were 61 and 63%, respectively. (B) A Bland-Altman plot of the data shown in (A). The vertical axis shows the difference in methylation values measured by the two methods (Mquant minus COBRA), whereas the horizontal axis shows the average methylation value measured by the two. The mean (SD) of the difference between methods was +1.6% (8.1%), indicating little evidence for bias between methods (P = 0.48). The center dashed horizontal line shows the mean difference, while the outside horizontal lines show the 95%LoAs (at −14.6%, +17.8%).
Both methods measured the midrange as well as extremes of methylation. On the three sites studied by both COBRA and Mquant, in vitro methylated DNA gave 90–100% methylation by both methods. Mquant measures of methylation in nine CpGs in the Avy allele of in vitro methylated DNA gave values from 90–98% with an average of 95 ± 2%. On the other extreme, we observed nine instances of CpG sites with <10% methylation by both COBRA and Mquant.
DISCUSSION
We describe a method to quantify DNA methylation from BGS four-dye-trace electropherograms. This method uses data from the thymine trace almost exclusively and thus avoids any complications due to independent normalization of A, G, C and T peaks in four-dye-trace electropherograms (16). This method uses available, established software to read electropherograms (Phred and Bioedit).
Our method analyzes sites independently and uses the same number of non-CpG Ts on either side of the analyzed site. We first attempted a similar quantification using a mean of most or all non-CpG Ts in electropherograms, but this gave poor results, and simple inspection of the numbers in the Phred output revealed that the mean was less than most of the non-CpG Ts early in the electropherogram and that the mean was greater than nearly all of the non-CpG T's late in the electropherogram (data not shown). The areas (and heights) of thymine peaks gradually decline over most electropherograms and this is likely responsible for this effect. By taking the same number of Ts on either side of the analyzed CpG an effect of this gradual peak area decline over the electropherogram is obviated. The peak areas (and heights) also vary locally so that it is important to average two or more to get a value for 100% T. Fitting methods such as linear regression on neighboring T peaks could probably be used with similar effect.
Any CpG site can have between 0 and 100% methylation. In the TCGA (TaqalphaI) site COBRA and Mquant differences are statistically significant yet the mean values are still within 10% methylation of each other. One possible explanation involves consistent differences readily observed in T peak areas and heights in electropherograms. Certain patterns, such as three successive Ts in a particular part of a sequence showing a gradual decline in height, are reproducible in multiple sequencings. This indicates that if a particular T derived from the C of a CpG is consistently above average height (and area) in its region of the sequence, the average methylation level measured by Mquant will be low. Similarly, if the particular T were consistently below average height (and area) in its region the average methylation level measured by Mquant would be high. Fortunately, differences that may be attributable to this effect at the TaqalphaI site are small. The ACGT (HpyCH4IV) site gave nearly identical mean methylation values by COBRA and Mquant as did the GCGG (AciI) site. These mean values were within 2% methylation of each other.
We agree with other groups (12,16) that BGS probably has limitations to quantification due to incomplete C-to-U conversion and imperfect specificity for only unmethylated Cs. However, we find good agreement with the established COBRA method when we quantify methylation in bisulfite electropherograms by averaging Ts on both sides of the target CpG and subtracting the T signal at the CpG site from this average T. To measure very high levels of methylation, e.g. 90–100%, it is necessary that the signal-to-noise ratio be very high, so that the average non-CpG T value is high and the noise at the CpG site is very low. For example, a noise level of 10% in the T trace at a CpG site leaves the maximum methylation detectable at 90% even if the DNA is actually 100% methylated. We made estimates of likely conversion levels and find them to be high (93–100%). Noise levels in sequencing and PCR may be higher than C signals due to incomplete C to T conversion.
Because it relies on nearby Ts to calculate methylation levels at CpGs, the Mquant method may not work as well in parts of some dense CpG islands, such as those of tumor suppressor genes, where there are few (or no) nearby Ts. In contrast, COBRA and some other methods would not be expected to show effects of nearby T density and thus COBRA may be a method of choice for such sequences. Mquant also relies on a high bisulfite conversion level and high-quality sequencing traces which may not always be available. For example, the algorithm of Lewin et al. (16) corrects for bisulfite conversion levels and aligns sequences and thus can work with sequencing traces that may not be useful with Mquant.
Mquant has several advantages over COBRA. Mquant quantifies methylation on multiple independent CpG sites from analysis of a single sequencing run. In contrast, COBRA usually analyzes the methylation of just one CpG site at a time. In most sequences only a minority of CpG sites are part of a restriction site as required for COBRA analysis. Mquant is amenable to robotic or manual high-throughput methods. For example, most or all of the steps could be done in a 96-well format up until the sequencing capillary electrophoresis which is also available in a 96 capillary format. In contrast, COBRA is generally done by gel electrophoresis with manual imaging and analysis of gels. These COBRA methods are laborious and, on average, probably less reliable than automated sequencing by capillary electrophoresis and fluorescent dye detection. Mquant also uses less than half of the DNA needed for one COBRA analysis.
Mquant works well over the entire range of methylation levels making it suitable for analyses of hypo- and hypermethylation and of methylation associated with imprinting. We obtained good overall correlations between Mquant and COBRA including levels near 0% and 100% methylation. Mquant and COBRA both gave high methylation levels (90–98%) with in vitro methylated DNA. There are always trace values for all four bases (including T) in electropherograms and there is always background noise in COBRA gel scans. Thus no measures gave a value of 100%.
Our use of the thymine trace data to quantify methylation has similarities to the method of Dikow et al. (12), but also differs in several ways. Like Mquant the Dikow algorithm can quantify from just the T trace values. However, the Dikow algorithm uses all Ts in a region without a way of choosing those well suited to quantify at a particular CpG site. In other words, they choose one set of Ts for quantification at multiple CpG sites whereas Mquant uses sets of Ts tailored to each CpG site. In Mquant each CpG uses a different set of Ts from every other CpG (unless two or more CpGs happen to have no Ts between them). Dikow et al. read electropherograms with Applied Biosystems Genescan software, whereas we used Phred and Bioedit. Dikow et al. report their maximum mean signal to be just over 80% methylation in DNA where they measured nearly 100% methylation by their established methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA) method. As discussed above, we are able to read methylation levels of 90–98% from in vitro methylated DNA.
Lewin et al. (16) use both the C and T trace values in a sophisticated but complex algorithm that ultimately normalizes the C and T traces to each other. They then use both the C and T trace data to calculate the level of methylation at each CpG. The Lewin algorithm also corrects for bisulfite conversion levels and aligns sequences. In contrast, Mquant quantifies methylation levels using only the T trace and thus does not require normalization or alignment. However, we mainly use sequences that are well aligned and that have high bisulfite conversion levels. The Lewin algorithm correction and alignment features allow it to use sequences that we might reject for Mquant.
We tested the Mquant method using different numbers of surrounding nontarget Ts (2–20) on either side of the target T/CpG. We found few differences, although correlations with COBRA were slightly higher when using a larger number of Ts (6–20).
Most methods to quantify methylation target only a few sites, require laborious, specialized laboratory techniques, or require highly specialized instrumentation. We used a specialized laboratory technique, COBRA, on three short sequences to show a very high correlation with the Mquant method. This method uses widely available techniques and instrumentation and should be useful in many laboratories to quantify DNA methylation levels.
ACKNOWLEDGEMENTS
We thank Dr Phillip Green for the Phred program and Dr Alan Diekman for loan of the GeneAmp PCR system 2700 used in this study. CAC acknowledges U.S. patent no. 6541680 and U.S. patent application 10402704. This work was supported by National Institute on Aging/National Institutes of Health [P01AG20641]; National Institute on Alcohol Abuse and Alcoholism/National Institutes of Health [R01AA016676]; Arkansas Biosciences Institute (Arkansas Tobacco Settlement Fund). Funding to pay the Open Access publication charges for this article was provided by National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Cooney CA. Epigenetics–DNA-based mirror of our environment? Dis. Markers. 2007;23:121–137. doi: 10.1155/2007/394034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu L, van Groen T, Kadish I, Tollefsbol TO. DNA methylation impacts on learning and memory in aging. Neurobiol. Aging, 2007 doi: 10.1016/j.neurobiolaging.2007.07.020. doi:10.1016/j.neurobiolaging.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ptak C, Petronis A. Epigenetics and complex disease: from etiology to new therapeutics. Annu. Rev. Pharmacol. Toxicol. 2008;48:257–276. doi: 10.1146/annurev.pharmtox.48.113006.094731. [DOI] [PubMed] [Google Scholar]
- 5.Robertson KD. DNA methylation and human disease. Nat. Rev. Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 6.Siegmund KD, Connor CM, Campan M, Long TI, Weisenberger DJ, Biniszkiewicz D, Jaenisch R, Laird PW, Akbarian S. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS. ONE. 2007;2:e895. doi: 10.1371/journal.pone.0000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szyf M, Weaver I, Meaney M. Maternal care, the epigenome and phenotypic differences in behavior. Reprod. Toxicol. 2007;24:9–19. doi: 10.1016/j.reprotox.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 8.van Vliet J, Oates NA, Whitelaw E. Epigenetic mechanisms in the context of complex diseases. Cell. Mol. Life Sci. 2007;64:1531–1538. doi: 10.1007/s00018-007-6526-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35:146–150. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- 11.Cooney CA, Eykholt RL, Bradbury EM. Methylation is co-ordinated on the putative replication origins of Physarum ribosomal DNA. J. Mol. Biol. 1988;204:889–901. doi: 10.1016/0022-2836(88)90049-6. [DOI] [PubMed] [Google Scholar]
- 12.Dikow N, Nygren AO, Schouten JP, Hartmann C, Kramer N, Janssen B, Zschocke J. Quantification of the methylation status of the PWS/AS imprinted region: comparison of two approaches based on bisulfite sequencing and methylation-sensitive MLPA. Mol. Cell Probes. 2007;21:208–215. doi: 10.1016/j.mcp.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ. Health Perspect. 2006;114:567–572. doi: 10.1289/ehp.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaminsky ZA, Assadzadeh A, Flanagan J, Petronis A. Single nucleotide extension technology for quantitative site-specific evaluation of metC/C in GC-rich regions. Nucleic Acids Res. 2005;33:e95. doi: 10.1093/nar/gni094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurmasheva RT, Peterson CA, Parham DM, Chen B, McDonald RE, Cooney CA. Upstream CpG island methylation of the PAX3 gene in human rhabdomyosarcomas. Pediatr. Blood Cancer. 2005;44:328–337. doi: 10.1002/pbc.20285. [DOI] [PubMed] [Google Scholar]
- 16.Lewin J, Schmitt AO, Adorjan P, Hildmann T, Piepenbrock C. Quantitative DNA methylation analysis based on four-dye trace data from direct sequencing of PCR amplificates. Bioinformatics. 2004;20:3005–3012. doi: 10.1093/bioinformatics/bth346. [DOI] [PubMed] [Google Scholar]
- 17.Nygren AO, Ameziane N, Duarte HM, Vijzelaar RN, Waisfisz Q, Hess CJ, Schouten JP, Errami A. Methylation-specific MLPA (MS-MLPA): simultaneous detection of CpG methylation and copy number changes of up to 40 sequences. Nucleic Acids Res. 2005;33:e128. doi: 10.1093/nar/gni127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfeifer GP, Riggs AD. Genomic sequencing by ligation-mediated PCR. Mol. Biotechnol. 1996;5:281–288. doi: 10.1007/BF02900367. [DOI] [PubMed] [Google Scholar]
- 19.Tost J, Dunker J, Gut IG. Analysis and quantification of multiple methylation variable positions in CpG islands by Pyrosequencing. Biotechniques. 2003;35:152–156. doi: 10.2144/03351md02. [DOI] [PubMed] [Google Scholar]
- 20.Uhlmann K, Brinckmann A, Toliat MR, Ritter H, Nurnberg P. Evaluation of a potential epigenetic biomarker by quantitative methyl-single nucleotide polymorphism analysis. Electrophoresis. 2002;23:4072–4079. doi: 10.1002/elps.200290023. [DOI] [PubMed] [Google Scholar]
- 21.Wong IH. Qualitative and quantitative polymerase chain reaction-based methods for DNA methylation analyses. Methods Mol. Biol. 2006;336:33–43. doi: 10.1385/1-59745-074-X:33. [DOI] [PubMed] [Google Scholar]
- 22.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clark SJ, Statham A, Stirzaker C, Molloy PL, Frommer M. DNA methylation: bisulphite modification and analysis. Nat. Protoc. 2006;1:2353–2364. doi: 10.1038/nprot.2006.324. [DOI] [PubMed] [Google Scholar]
- 25.Carr IM, Valleley EM, Cordery SF, Markham AF, Bonthron DT. Sequence analysis and editing for bisulphite genomic sequencing projects. Nucleic Acids Res. 2007;35:e79. doi: 10.1093/nar/gkm330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis TL, Trasler JM, Moss SB, Yang GJ, Bartolomei MS. Acquisition of the H19 methylation imprint occurs differentially on the parental alleles during spermatogenesis. Genomics. 1999;58:18–28. doi: 10.1006/geno.1999.5813. [DOI] [PubMed] [Google Scholar]
- 27.Lucifero D, Mertineit C, Clarke HJ, Bestor TH, Trasler JM. Methylation dynamics of imprinted genes in mouse germ cells. Genomics. 2002;79:530–538. doi: 10.1006/geno.2002.6732. [DOI] [PubMed] [Google Scholar]
- 28.Cooney CA, Dave AA, Wolff GL. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J. Nutr. 2002;132:2393S–2400S. doi: 10.1093/jn/132.8.2393S. [DOI] [PubMed] [Google Scholar]
- 29.Rakyan VK, Chong S, Champ ME, Cuthbert PC, Morgan HD, Luu KVK, Whitelaw E. Transgenerational inheritance of epigenetic states at the murine AxinFu allele occurs after maternal and paternal transmission. Proc. Natl Acad. Sci. USA. 2003;100:2538–2543. doi: 10.1073/pnas.0436776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 31.Rudi K, Rud I, Holck A. A novel multiplex quantitative DNA array based PCR (MQDA-PCR) for quantification of transgenic maize in food and feed. Nucleic Acids Res. 2003;31:e62. doi: 10.1093/nar/gng061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 33.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 34.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1:307–310. [PubMed] [Google Scholar]
- 35.Bland JM, Altman DG. Measuring agreement in method comparison studies. Stat. Methods Med. Res. 1999;8:135–160. doi: 10.1177/096228029900800204. [DOI] [PubMed] [Google Scholar]
- 36.Qiu P, Soder GJ, Sanfiorenzo VJ, Wang L, Greene JR, Fritz MA, Cai XY. Quantification of single nucleotide polymorphisms by automated DNA sequencing. Biochem. Biophys. Res. Commun. 2003;309:331–338. doi: 10.1016/j.bbrc.2003.08.008. [DOI] [PubMed] [Google Scholar]