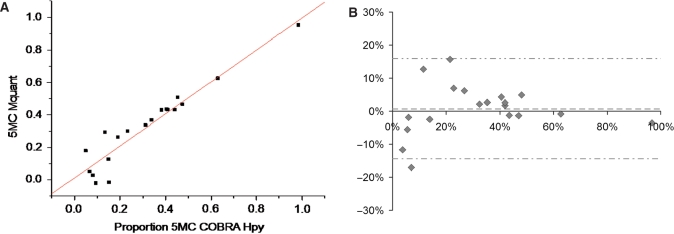
Figure 2.
(A) A plot of the percent DNA methylation on the CpG of an ACGT site from 19 different DNA samples determined by COBRA versus the percent methylation on this same site determined by peak areas on forward sequence electropherograms. The results of the two methods were highly correlated (R = 0.95, P < 10−9) and in good agreement (estimate ± standard error = 0.98 ± 0.079 for slope, and 0.012 ± 0.031 for y-intercept). The average percent methylation measured by COBRA and Mquant were both 32%. (B) A Bland-Altman plot of the data shown in (A). The vertical axis shows the difference in methylation values measured by the two methods (Mquant minus COBRA), whereas the horizontal axis shows the average methylation value measured by the two. The mean (SD) of the difference between methods was +0.72% (7.6%), indicating little evidence for bias between methods (P = 0.68). The center dashed horizontal line shows the mean difference, while the outside horizontal lines show the 95%LoAs (at −14.4%, +15.9%).