Abstract
Single amino acid changes at specific DNA contacts of histones H3 and H4 generate SWI/SNF-independent (Sin) mutants in yeast. We have analyzed the effect of the Sin mutation at R45 of histone H4 on cell survival following UV irradiation, nucleotide excision repair (NER) and chromatin structure. We find that this mutation renders yeast cells more resistant to UV damage and enhances NER at specific chromatin loci. In the transcriptionally silent HML, repressed GAL10 and the constitutively active RPB2 loci, H4 R45 mutants exhibit enhanced repair of UV-induced cyclobutane pyrimidine dimers (CPDs) compared to wild-type (wt). However, the H4 R45 mutation does not increase the transcription of NER genes, disrupt transcriptional silencing of the HML locus or alter repression in the GAL10 locus. We have further shown that the H4 R45C mutation increases the accessibility of nucleosome DNA in chromatin to exogenous nucleases and may expedite nucleosome rearrangements during NER. Taken together, our results indicate that the increased repair observed in Sin mutants is a direct effect of the altered chromatin landscape caused by the mutation, suggesting that such subtle changes in the conserved histone residues can influence the accessibility of DNA repair factors in chromatin.
INTRODUCTION
Cellular DNA is continually exposed to threats from various endogenous and exogenous sources of damage that can compromise the functional integrity of the genome. Assaults caused by different types of genotoxic agents exert evolutionary pressure on all organisms, which have developed sophisticated responses to cope and survive (1). An important form of damage response exhibited by cells is their ability to repair DNA lesions caused by environmental sources such as UV radiation, ionizing radiation and chemical agents (2). Failure to repair such DNA lesions is known to cause mutations that can lead to genomic instability, cell death or life-threatening diseases like cancer in higher eukaryotes.
Nucleotide excision repair (NER) is an important mechanism for removal of a wide variety of bulky DNA lesions, such as cis-syn cyclobutane pyrimidine dimers (CPDs) and (6–4) photoproducts caused by ultraviolet (UV) radiation. NER consists of two pathways: global genome repair (GGR) and transcription-coupled repair (TCR) (3–5). TCR is specifically responsible for repair of the transcribed strand of actively transcribed genes and appears to be triggered by stalled elongating RNA polymerase. GGR is involved in removal of lesions from unexpressed regions of the genome and the nontranscribed strand of actively transcribed genes. In eukaryotic cells, accessibility of lesions in highly condensed 100–400 nm chromatin fibers adds further complexity (6,7) and the compact DNA packaging into chromatin is refractory to DNA repair enzymes (8,9). This structural hierarchy hinders the access of nuclear factors that detect and subsequently repair lesions in nuclear DNA. However, the chromatin environment is dynamic and early studies showed that chromatin rearrangements occur during NER in intact cells (9,10).
Nucleosomes, the basic structural component of chromatin represents the first level of DNA compaction in eukaryotes (11). This fundamental unit consists of 147 bp of DNA wrapped in 1.65 superhelical turns around an octamer of the core histones H2A, H2B, H3 and H4. The histone folds of H3 and H4, as well as H2A and H2B, interlock to form a stable H3–H4 tetramer and two H2A–2B dimers, giving rise to the canonical core octamer assembly (12). Interaction is primarily between amino acids in the α-helices or intervening loop regions of the core histones and the phosphodiester backbone of the DNA. Of the 14 sites where the DNA minor groove contacts the histone octamer, 12 are penetrated by side-chains of an arginine residue located in either a histone fold domain or an N-terminal tail region (11). Furthermore, at 8 of these 12 sites, a threonine side chain from a neighboring histone forms either direct (H3 T118 with H4 R45) or water-mediated hydrogen bonds with the penetrating arginine residue (13).
Mutations that alter one of a cluster of amino acid residues lying in the specific DNA-histone contact sites of histones H3 and H4 are known to generate a class of mutants called SWI/SNF-independent (Sin) mutants (14). These mutants were identified by their ability to carry on transcription in the absence of yeast chromatin remodeling complex SWI–SNF. The Sin mutations are found to be clustered in the L1L2 loop regions of the H3–H4 tetramer that participates in binding the central two turns of nucleosomal DNA (14), and broadly fall into two groups: (i) mutations that target H4 R45, H3 T118 and H3 R116 are located very near the dyad center contact points (15) and (ii) mutations that target H4 V43 or H3 E105, are located more distant from the dyad center. Previous studies demonstrated that Sin mutations do not alter the histone octamer stoichiometry, but in general render nucleosome DNA more accessible to enzymes such as micrococcal nuclease and Dam methylase (16–18). Sin mutant nucleosomes mobilize at lower temperatures and dissociate at lower salt concentrations (14,19), with an increased ability to reposition on the DNA. Interestingly, nucleosome arrays reconstituted with octamers containing the H4 R45C mutant histone exhibit a defect in magnesium-dependent intramolecular folding (18).
The Arg 45 residue of histone H4 that has been mutated in our study is one of the 10 Arg residues whose side chain protrudes into the minor groove of the DNA (11,13). Mutation of H4 R45 to cysteine or histidine results in an ‘empty’ minor groove, leading to a disruption of the histone–DNA interactions. However, this mutation has only a moderate effect on global nucleosome structure (14). It is suggested that the H4 R45 mutation leads to an altered nucleosome state that mimics SWI/SNF-dependent nucleosome disruption, a typical consequence of Sin mutations. In the present study, we examined the effect of H4 R45C and R45H Sin mutations on NER of UV damage. Our results show that cells containing these versions of histone H4 have more efficient NER than wild-type (wt) cells, and are more resistant to UV-induced cell death. In agreement with past reports (16,17), we find increased accessibility of nucleosomal DNA in the chromatin of these cells. These results indicate that the subtle change in chromatin induced by a single-site mutation in a core histone of yeast cells allows enhanced UV survival and increased DNA repair in specific chromatin regions.
MATERIALS AND METHODS
Construction of H4 R45 Sin histone mutants
Plasmid shuffle method was used to introduce mutated versions of histone H4 gene. Yeast strain WY121, a derivative of W303, in which all four histone H3 and H4 genes have been disrupted was used. WY121 bears plasmid pJL001 (CEN URA3 HHT2-HHF2) containing a single copy of the wt histone gene pairs HHT2-HHF2 and a counter-selectable marker URA3 (20).
Microarray data accession number
The data discussed in this report have been deposited in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and is accessible through GEO series accession number GSE11282. The strain was transformed with plasmids bearing either H4 R45C or R45H (HHT2-hhf2) derivative of HHT2-HHF2 plasmid pJW028 (CEN ADE2 HHT2-HHF2) having a selectable marker ADE2. The QuickChange Kit (Stratagene, La Jolla, CA, USA) was used to introduce the R45C/R45H mutation into plasmid pJW028. It was found that the yeast strain transformed with H4 R45H plasmid bore single plasmid population expressing the mutant H4 gene. The H4 R45C cells, however, contained both wt (HHT2-HHF2) and the mutant H4 R45C (HHT2-hhf2) expressing plasmids. As already reported (21), it is possible that, in this wt background yeast strains bearing H4 R45C as the sole source of histone H4 are not viable.
UV-sensitivity and repair assays
Yeast cells were grown at 30°C in YPD to early log phase (A600 ∼ 0.6), harvested, placed on ice for 1 h, and washed with ice-cold PBS (phosphate-buffered saline). The washed cells were resuspended in ice-cold PBS and irradiated with 100 J/m2 UV light (254 nm). The cells were then allowed to repair by incubation in prewarmed YPD for various time periods in dark at 30°C followed by pelleting and DNA isolation using the glass-bead method as described previously (22). For UV sensitivity assay, cells were diluted to different concentrations, spread on YPD plates and irradiated with the indicated UV doses. Colonies were counted after 48 h of incubation at 30°C in the dark.
Locus-specific repair analyses
NruI, Bsp1286I and EcoRI–EcoRV were used to release the 3.4-kb fragment containing the RPB2 gene, the 2.3-kb fragment from the HML locus and the 2.2-kb fragment of the GAL10 locus, respectively. The number of CPDs in specific restriction fragments was determined as described previously (23,24). Briefly, equal amounts of purified and restriction-digested DNA were either mock treated or treated with T4 endonuclease V for 60 min at 37°C. DNA was denatured and electrophoresed on 1% alkaline agarose gel, transferred to Hybond N+ membranes, and hybridized with specific radioactive DNA or RNA probes. The probes for RPB2 locus were generated using linearized plasmid pKS212 (3). For the HML locus, a fragment representing nucleotides +1 to +518 was generated from the HMLα1 ORF by PCR amplification. For the GAL10 probe a fragment representing +1 to +800 bp relative to the start site was used. The fragments were radiolabeled using [α32P]dATP and Prime-It Random Primer Kit (Stratagene). Southern blots were quantified using PhosphorImager and IMAGE-QUANT software (GE Healthcare, Piscataway, NJ, USA). The level of repair was calculated as the amount of CPDs remaining per fragment, according to Poisson distribution (3).
Chromatin accessibility assay
Micrococcal nuclease (MNase) digestion was done following protocols as described (25,26). Briefly, 100 ml of mid-log phase (∼1.0 –2.0 × 107 cells/ml) yeast cells were pelleted, washed with 1 M sorbitol, suspended in YLE (10 mg/ml zymolyase in 1 M sorbitol and 5 mM β-mercaptoethanol) and incubated at 22°C for 30 min. Cells were repelleted and washed twice with sorbitol wash buffer (1 M sorbitol, 1 mM PMSF, 2 mM β-mercaptoethanol). Sphaeroplasts were then suspended in sphaeroplast digestion buffer (1 M sorbitol, 50 mM NaCl, 10 mM Tris–HCl, pH 7.5, 5 mM MgCl2, 1 mM CaCl2, 1 mM β-mercaptoethanol, 0.5 mM spermidine and 0.075% v/v NP-40), divided into 200-μl aliquots and digested with varying concentrations of MNase (10 U/μl) for 10 min at 37°C. The reactions were terminated with 0.1 vol of stop solution (5% SDS, 250 mM EDTA) followed by proteinase-K treatment for 2 h at 55°C. Samples were extracted twice with phenol:chloroform, treated with 5 μg/ml of RNase A and ethanol precipitated. DNA was resuspended in TE (10 mM Tris pH-8.0, 1 mM EDTA) and electrophoresed on 1.2% agarose gel. Southern blots were done with random primed 32P-labeled probes specific for HML or RPB2 locus.
For restriction enzymes accessibility assay, log-phase yeast cells were treated with 100 J/m2 UV light followed by repair incubation for various time periods. Nuclei were isolated as described above and subjected to EcoRV digestion for 20 min. DNA was purified, further digested with Bsp1286I to release the HML fragment and resolved on 1% agarose gel. Southern blot analysis was done using random primed 32P-labeled 150-bp fragment spanning +1 to +150 nt of HMLα1 ORF.
Genome-wide expression profiling
For each genome-wide expression experiment, three mutant and three wt yeast cultures were grown in YPD medium to a final optical density at 600 nm of 0.4–0.7 and then harvested. Total RNA was isolated from each yeast culture and used to prepare cDNA and biotinylated c-RNA, as described previously (20). The cRNA was then hybridized to a single S98 genome oligonucleotide array and scanned following standard protocols (Affymetrix). Intensities were captured using GeneChip software (Affymetrix), and a single raw expression level for each gene was determined.
Data analysis
The data from each chip were normalized using GeneChip software (version 5; Affymetrix). The microarray data were analyzed using modified triple error model to calculate the significance (P-value) of the observed change in mRNA levels. A change in mRNA levels was deemed significant based on the following criteria: (i) the average change up or down was >2-fold; (ii) the change (up or down) in each replicate experiment was >1.5-fold and (iii) the absolute intensity change was above background levels. [For details, see the report by Martin et al. (20)].
RT–PCR analysis
Cells were grown to log phase under the same conditions as used for microarray experiments. Total RNA was isolated from each yeast culture and 5 μg of RNA from each sample was reverse transcribed using Superscript III RT enzyme (Invitrogen, Carlsbad, CA, USA), as per manufacturer's instructions. The product was treated with RNase H and PCR amplified for 25 cycles using gene-specific primers.
RESULTS
The H4 R45 mutants used in this study were generated by plasmid shuffling into a wt yeast strain, where the four chromosomal copies of H3 and H4 histone genes have been disrupted, and the wt or mutant H3 and H4 genes are expressed on a plasmid (see Materials and methods section). As mentioned earlier, unlike the H4 R45H mutant strain, we were unable to generate an H4 R45C strain whose sole source of histone H4 was the mutant plasmid. Indeed, we examined over 20 different H4 R45C mutant clones, by sequencing the isolated plasmid, and in each case at least a small amount of wt plasmid was present (unpublished data). This result suggests that the H4 R45C mutant strain can not survive with H4 R45C as the sole source of histone H4, and is in agreement with past reports on this mutant (21). However, Sin mutants are known to be partially dominant to wt histone genes and the mutant histones lead to a Sin phenotype even in the presence of wt histones (15).
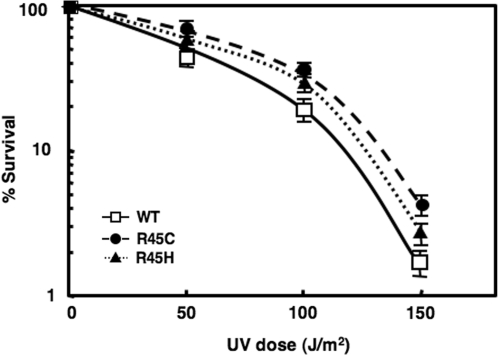
Both the H4 R45 mutants used in our experiments exhibit a slower growth rate at 30°C, compared to wt cells, and exhibit a slightly more resistant (lower cell death) phenotype to UV irradiation (Figure 1). Interestingly, among several other Sin histone mutants (H3 R116H, H3 T118I, H4 V43I) that we tested, only H4 R45 mutants showed resistance to UV irradiation compared to wt (Figure 1 and data not shown). These observations led us to explore DNA repair of UV damage as a possible defence mechanism affected by such a subtle change in chromatin.
Figure 1.
UV sensitivity of wt, H4 R45C and H4 R45H. Colony-forming ability following UV irradiation was monitored in exponentially growing cultures. Cells were appropriately diluted, spread on YPD plates, subjected to the UV doses shown and their survival monitored. For each strain, data represent the mean ± 1 SD for four independent experiments.
H4 R45 mutants have enhanced NER activity at specific chromatin loci
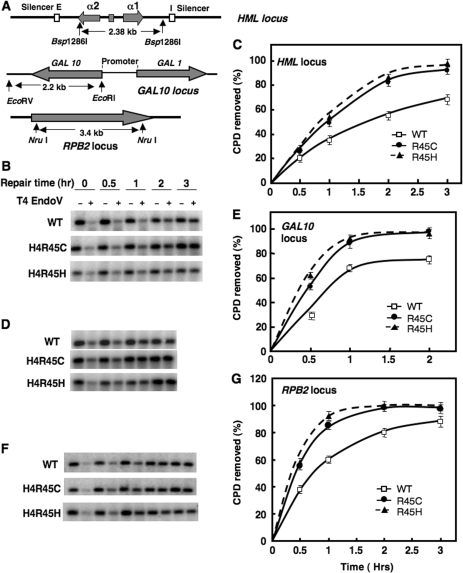
For NER studies, yeast cells were irradiated with 254-nm UV light (100 J/m2) and incubated for different time periods following irradiation. Based on the chromatin structure, three different kinds of loci in yeast were tested for CPD removal: (i) the nucleosome-loaded and silenced mating-type locus HML; (ii) the nucleosome-loaded inducible GAL10 locus under transcriptionally repressed conditions; and (iii) the constitutively expressed RPB2 gene, encoding the second largest subunit of RNA Pol II. Total DNA was isolated, digested with appropriate restriction enzymes and cut specifically at CPDs with T4 endonucleaseV (T4 endoV). Repair of CPDs was analyzed following separation of the cleaved DNA fragments on alkaline agarose gels as previously described (3).
Repair of CPDs in the HML locus of H4 R45 mutant and wt cells was monitored in a 2.38-kb Bsp1286I fragment (Figure 2A). This fragment contains several positioned nucleosomes (27) and bound silencing proteins Sir2, Sir3 and Sir4 which form the Sir complex (28). As shown in Figure 2B and C, CPD removal is about 50% more efficient in H4 R45 mutant cells than wt after 2 h of repair, and is essentially complete by 3 h after irradiation (compared to only ∼70% repair in wt cells). Similar results were obtained for CPD repair in a 2.2-kb EcoRI–EcoRV fragment containing the GAL10 gene (Figure 2A). Under repressed conditions (i.e. in glucose medium), this region contains nucleosomes of varying stability (29) and no bound silencing complex (30). As with the HML locus, we found that both of the H4 R45 mutants have more rapid NER in the GAL10 locus, where ∼40% more CPDs are removed by 1 h after irradiation compared to wt cells (Figure 2D and E).
Figure 2.
Nucleotide excision repair in the HML, GAL 10 and RPB2 loci of H4 R45 cells. Diagram of the HML, GAL10 and RPB2 loci are shown in (A), where pertinent restriction sites and fragment lengths are denoted. For repair experiments, cells were UV irradiated (100 J/m2) and incubated in the dark for various time periods (0.5 to 3 h). Genomic DNA was isolated, digested with appropriate restriction enzyme and then digested to completion with T4 endo V. Southern analyses were performed using radiolabeled probes specific for HML, GAL10 and RPB2 loci, respectively. Representative alkaline agarose gels showing CPD removal from the 2.3-kb Bsp1286I fragment (HML), 2.2- kb EcoRI–EcoRV fragment (GAL10) and the 3.4-kb NruI fragment (RPB2) are shown in (B), (D) and (F), respectively. The time courses calculated for CPD removal from the HML, GAL10 and RPB2 loci are shown in (C), (E) and (G), respectively. For each strain, data represent the mean ± 1 SD for three independent experiments.
We also examined removal of CPDs from a 3.4-kb NruI fragment in the RPB2 locus (Figure 2A). Once again, it was found that NER of UV damage in both H4 R45 mutants is more rapid than in wt cells (Figure 2F and G). Analogous to the GAL10 locus, following 1 h repair incubation, both H4 R45C and R45H cells show ∼40% more efficient CPD removal from the RPB2 locus, compared to wt. Overall, our results indicate that the two H4 R45 mutant strains have increased resistance to UV by virtue of increased repair of UV-induced DNA damage.
Chromatin in H4 R45C cells has increased accessibility to nucleases
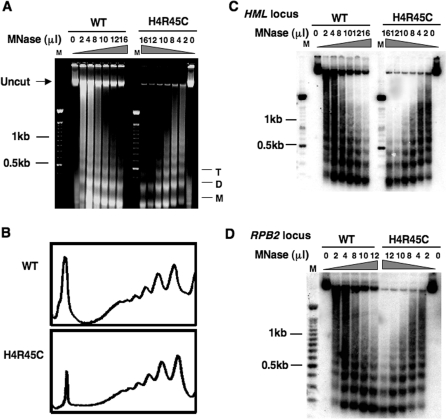
It was reported previously that H4 R45 mutant chromatin has increased nuclease sensitivity compared to wt (17). As such a feature could influence NER, we examined the MNase accessibility of DNA in chromatin from the H4 R45C cells used in our experiments and compared this to wt chromatin. We investigated the DNA accessibility of both bulk chromatin and the three different chromatin loci tested for NER activity (Figure 2).
For these studies, spheroplasts were isolated from wt and H4 R45C cells and treated with varying concentrations of nuclease. Comparison of the MNase digestions show that the H4 R45C chromatin is more sensitive compared to wt. As shown in Figure 3A, the same amount of enzyme generated nucleosome ladders of 3–7 repeats in wt chromatin compared to only di- or tri-nucleosome repeats in the mutant chromatin (e.g. compare 10, 12 and 16 units lanes). Moreover, while nearly equal amounts of DNA were loaded on each agarose gel (quantified from the total DNA in undigested samples), MNase digestion always yielded more digested material in mutant samples compared to wt (Figure 3A, uncut band). Scans of lanes with equivalent digestion conditions, however, revealed no distinguishable differences between the nucleosome repeats of H4 R45C and wt cells (Figure 3B), indicating that similar nucleosome loading occurs in both wt and mutant chromatin. Moreover, Southern blot analyses with HML, RPB2 or GAL10 probes indicate that MNase has increased accessibility to each of these loci in H4 R45C chromatin (Figure 3C and D, data not shown). The H4 R45C mutation, therefore, does not appear to alter nucleosome spacing, yet renders chromatin more accessible to MNase. These observations may reflect the increased ability of Sin histone octamers to reposition (or slide) on DNA with elevated temperature and/or disrupt higher order folding of chromatin, as reported earlier (14,18).
Figure 3.
Accessibility of H4 R45C chromatin to MNase digestion. Isolated spheroplasts from wt and H4 R45C cells were treated with different concentrations of MNase, as described in Materials and methods section, and genomic DNA was isolated and separated on 1.2% agarose gels. (A) Digestion profile of bulk chromatin, stained with ethidium bromide. M, D and T denote mononucleosome, dinucleosome and trinucleosome DNA, respectively. (B) Scans of 10 μl of MNase (10 U/μl) lanes shown in (A). Southern blot hybridization with probe to the (C) HML locus and (D) RPB2 locus.
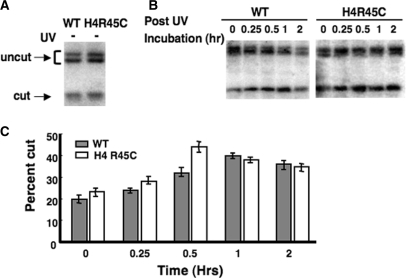
To further explore the accessibility of DNA in H4 R45 mutant chromatin, we also examined accessibility of the lone EcoRV restriction site, located within a positioned nucleosome, in the HML locus (27). As shown in Figure 4A, only a minor fraction (∼20%) of the EcoRV sites are cleaved in either wt or H4 R45C nuclei from unirradiated cells. However, this fraction increases by more than 2-fold during the repair process following UV irradiation (Figure 4B). Interestingly, a consistent difference was observed between the cell types in the maximum fraction of cut sites during repair. The maximum fraction cut (∼45%) was observed after 30 min of repair incubation in H4 R45C nuclei, while the maximum for wt nuclei (∼40%) was observed after 1 h of repair incubation (Figure 4B and C). Finally, in both strains, this enhanced EcoRV accessibility started to decline after 1 h of repair incubation (Figure 4C). These results support the ‘unfolding-refolding’ model for NER of UV damage (31), and again may reflect enhanced DNA accessibility in H4 R45 mutant chromatin.
Figure 4.
Accessibility of the EcoRV site in HML chromatin. Nuclei were isolated from wt and H4 R45C cells (A) before or (B) after UV irradiation (100 J/m2) and digested with EcoRV, as described in Materials and methods section. Arrows show the positions of the uncut and cut bands. (C) Percent of HML DNA in chromatin accessible to EcoRV, where bars show values obtained for wt (shaded) and H4 R45C (open) cells. Data represent the mean ± 1 SD of three independent experiments.
H4 R45 mutations increase RPB2 expression without affecting HML silencing
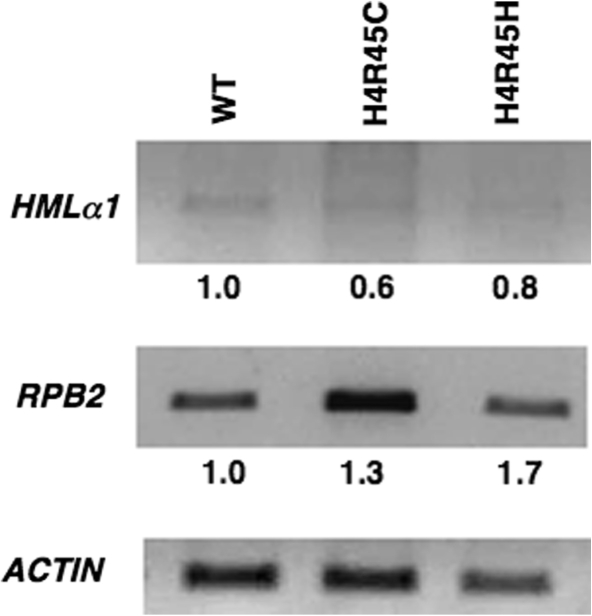
It is possible that H4 R45 mutations alter the transcription levels of the genes we tested for NER activity. Therefore, RT-PCR analyses were performed on total RNA isolated from H4 R45 mutants using gene specific primers. We observed a small increase of 1.3- and 1.7-fold in the level of RPB2 transcripts for H4 R45C and H4 R45H cells, respectively, when compared to wt (Figure 5, middle panel). However, no significant amount of HML α1 transcript was observed in either of the H4 R45 mutants compared to wt (Figure 5, top panel), indicating that these mutations do not affect transcriptional silencing in the HML locus. Similarly, no significant transcription of the GAL10 gene was found in any of these cells grown in glucose (data not shown). These results indicate that the increased repair of the HML and GAL10 loci do not reflect increased transcription in the H4 R45 mutant cells. On the other hand, the enhanced NER observed in the RPB2 gene, most likely reflects, at least in part, the increased transcription of RPB2 in H4 R45 mutant cells.
Figure 5.
RT–PCR analysis of transcription in H4 R45 mutants. RT–PCR analysis was performed on total RNA from wt, H4 R45C and H4 R45H cells using primers specific for the RPB2 and HML genes, as described in Materials and methods section. The RT–PCR product of ACTIN was used as a loading control to normalize the transcript levels of RPB2 and HMLα1 from each strain. The normalized value for the wt cells was set as 1.0.
Increased TCR in H4 R45C cells leads to faster repair in the RPB2 locus
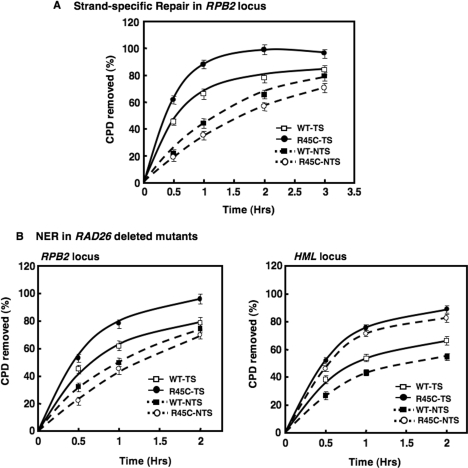
As transcription of the RPB2 gene was found to increase in the H4 R45 mutant cells, we examined TCR (i.e. strand-specific repair) in the RPB2 locus of the H4 R45C cells. As shown in Figure 6A, TCR in the RPB2 locus is more efficient in H4 R45C cells compared to wt. Over 95% of the lesions in H4 R45C cells are removed within 2 h of repair incubation, compared to <75% in wt cells. In contrast, CPD removal in the nontranscribed strand of RPB2 is slower in H4 R45C cells compared to wt (Figure 6A). These results suggest that faster repair of the RPB2 locus in H4 R45 mutant cells reflects increased TCR, while GGR is somewhat suppressed, possibly by high levels of TCR.
Figure 6.
Transcription coupled repair and effect of RAD26 deletion on NER of the RPB2 gene in H4 R45C cells. (A) Both wt and H4 R45C cells were treated, and samples prepared, as described in legend to Figure 2. Southern analyses were performed using radiolabeled RNA probes specific for transcribing and nontranscribing strands of RPB2. The time course for CPD removal from each strand of the 3.4-kb NruI fragment of RPB2 are shown. (B) CPD removal from the RPB2 and HML loci of H4 R45C and wt strains was examined in UV irradiated cells with and without RAD26, as described in Materials and methods section. The corresponding time courses for CPD removal from the 3.4-kb NruI fragment (RPB2) and 2.3-kb Bsp1286I fragment (HML) are shown. Data represent the mean ± 1 SD for three independent experiments.
Deletion of RAD26 reduces the rapid rate of repair at the RPB2 locus without affecting repair of the HMLlocus in H4 R45C cells
To confirm that the increased repair of the RPB2 gene in H4 R45C cells is due to enhanced TCR, the effect of RAD26 deletion on NER was examined. Inactivation of RAD26 in yeast is known to cause defective TCR of UV-damaged DNA (32). We found that repair of CPDs at the RPB2 locus decreases significantly in H4 R45CΔrad26 cells (Figure 6B). Reduced repair is also observed in wtΔrad26 cells compared to wt, but the relative decrease is much less than that observed for the same comparison for H4 R45C cells (Figure 6B, compare open and closed symbols for each strain). These results strongly suggest that TCR is primarily responsible for the enhanced repair of the transcriptionally active RPB2 locus in H4 R45C cells. Importantly, however, the RAD26 deletion has little effect on repair of the HML locus (Figure 6C). NER in the HML locus of H4 R45CΔrad26 cells remains more efficient than both wt and wtΔrad26 cells (Figure 6C, compare open and closed symbols for each strain). The RAD26 deletion, however, does yield a slight reduction in NER in each strain.
H4 R45 mutation does not affect expression of NER genes
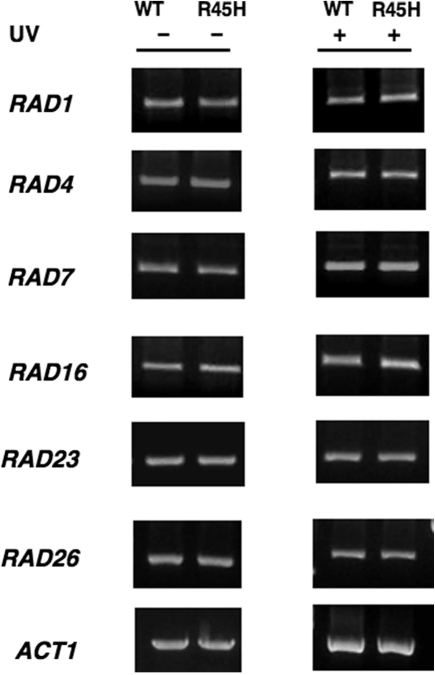
Another explanation for the enhanced NER in H4 R45 mutant cells is increased expression of NER genes. Therefore, we examined the change in mRNA levels using whole-genome Affymetrix oligonucleotide arrays. For each microarray experiment, triplicate mRNA samples from mutant and wt strains were profiled. A total of 475 genes were found to be upregulated (>2-fold increase) and 21 genes downregulated (>2-fold decrease) in the H4 R45H cells compared to wt. Among the upregulated genes is PHO5 (3.6-fold), which was previously reported to have disrupted repression in H4 R45 mutant cells (17). As shown in Table 1, among the commonly known NER genes only RAD2 was found to be increased (2.04-fold) in the mutant strain compared to wt. The microarray results were confirmed by RT-PCR experiments (Figure 7), performed on total RNA from wt and mutant cells treated with or without UV radiation. Thus, both assays indicate that the H4 R45 mutation causes little or no change in expression of the NER genes.
Table 1.
Comparative expression analysis of NER genes in H4 R45H and wt cells
| Gene name | Specific function | Increase (n-fold)a |
|---|---|---|
| RAD 1 | DNA endonuclease; DNA damage recognition | NS |
| RAD10 | DNA endonuclease; DNA damage recognition | NS |
| RAD14 | DNA endonuclease; DNA damage recognition | NS |
| RAD2 | DNA endonuclease | 2.0 |
| RAD4 | DNA damage binding | NS |
| RAD23 | DNA damage binding | NS |
| RAD26 | Functions in TCR | NS |
| RAD7 | DNA-dependent ATPase, DNA damage recognition | NS |
| RAD16 | DNA-dependent ATPase, DNA damage recognition | NS |
| PHO5 | Repressible acid phosphatase | 3.6 |
aNS, not significant [for details, see Materials and methods section, and Martin et al. (20)]
Figure 7.
RT–PCR analyses of expression of NER genes. RT–PCR analysis was performed on total RNA isolated from wt and H4 R45H cells, following treatment with or without 100 J/m2 UV radiation, using gene-specific primers, as described in Materials and methods section.
DISCUSSION
The structural hierarchy of chromatin plays an integral role in determining the distribution of damage within DNA, as well as, repair of this damage (8). For example, studies with Saccharomyces cerevisiae have revealed that both CPDs and 6-4PPs are removed more rapidly from nucleosome-free regions and linker DNA compared to nucleosome cores (33,34). In this report, we have shown the effect of Sin mutations R45C and R45H in histone H4 of yeast on cell survival, excision repair following UV damage, chromatin structure and transcription.
The H4 R45 mutation enhances repair in specific chromatin loci
Early experiments showed that nucleosome rearrangements occur during NER in human cells (10). These findings led to the ‘unfolding-refolding’ model of nucleosome rearrangement during NER (31,35), which has been given the designation ‘access, repair, restore’ (ARR) by Green and Almouzni (36). Recent findings show that the heterodimer Rad4-Rad23, interacts with the chromatin remodeling complex Swi/Snf during NER in intact yeast cells (37), indicating that rearrangement in at least some regions of chromatin is required for efficient NER. It has been reported that Sin histone mutants require less thermal energy for ‘nucleosome sliding’ along the DNA, which may reflect increased nucleosome mobility and disrupted higher order chromatin structure in vitro (14,18,19). Here we report that, H4 R45 mutant strains are more resistant to UV damage (Figure 1), which is unusual for yeast mutants (1). We examined removal of UV-induced CPDs in three different types of chromatin in yeast: transcriptionally silenced chromatin (HML locus) with positioned nucleosomes and bound Sir complexes (27,28); transcriptionally repressed (GAL10 locus) with nucleosomes of varying stability (29) and no bound silencing complexes (30,38); and transcriptionally active chromatin (RPB2 locus). We found that in each case NER is more rapid in H4 R45C and H4 R45H cells than in wt (Figure 2). Thus, our results suggest that the H4 R45 mutation increases the repair activity in specific chromatin loci in yeast.
H4 R45 mutant chromatin provides a better ‘landscape’ for NER
It has been hypothesized that mutation at R45 of histone H4 may weaken the central wrap of DNA around the histone octamer, leading to an altered nucleosome structure. Indeed, the H4 R45 mutations studied in this report, are known to (i) render the DNA more accessible to MNase and Dam methylase, (ii) eliminate Mg2+-dependent intramolecular folding of nucleosomal arrays and (iii) have a pronounced effect on thermally driven nucleosome sliding (14,17–19). Moreover, nucleosome arrays reconstituted with histone octamers containing H4 R45C or R45H mutations generate improved substrates for SWI/SNF remodeling complex (18). Therefore, a compelling argument is that chromatin in H4 R45 cells is a better ‘landscape’ for processing by repair proteins, and this accounts for the enhanced repair we observe in these cells.
In agreement with this hypothesis, our results show that DNA in bulk chromatin, as well as in HML, GAL10 and RPB2 loci, is more accessible to MNase in H4 R45C cells compared to wt (Figure 3 and data not shown). Furthermore, the MNase digestion patterns indicate that nucleosome spacing in these cells is similar to wt. As mentioned previously (18), this implies that H4 R45 mutant histones form nucleosomes similar to wt histones and the mutation does not alter nucleosome loading or spacing; and it is higher order folding of chromatin that is most likely affected in these cells.
To examine accessibility of nucleosome DNA in H4 R45C cells during NER, we monitored accessibility of the single EcoRV recognition site normally present within a positioned nucleosome in the HML locus. As shown previously for the repressed MFA2 locus (39), we observe that in both cell types the accessibility of the EcoRV cutting site increases after UV irradiation. Interestingly, in H4 R45C cells, the maximum fraction of these sites exposed is slightly higher and achieved at an earlier repair time (30 min) than in wt cells (1 h; Figure 4). These data further support the notion that the H4 R45 mutant chromatin is more accessible during NER than in wt, thereby improving the efficiency of DNA repair in chromatin.
Dissecting the effect of H4 R45 mutation on NER and transcription
It has been reported that the H4 R45 mutation leads to impaired nucleosome-mediated repression of the PHO5 gene (17,21). Therefore, we wondered if the enhanced repair rate observed in H4 R45 mutants is an indirect effect of enhanced transcription of the three loci tested. However, RT-PCR experiments showed that neither the transcriptional silencing of the HML locus nor the repression of the GAL10 gene is disrupted in H4 R45 mutant cells. On the other hand, expression of the RPB2 gene was found to be increased (1.3- to 1.7-fold) in the H4 R45 mutant cells compared to wt. Therefore, we examined strand-specific repair in this locus, and found that the enhanced repair is the result of more rapid repair of the transcribed strand (Figure 6). This demonstrated that TCR is the primary repair pathway for removal of CPD lesions from this locus, and the increased transcription rate leads to more efficient CPD removal in the RPB2 locus of H4 R45C cells. Interestingly, repair in the nontranscribed strand of RPB2 in H4 R45C cells occurs at a slower rate than either TCR or GGR in wt cells. This correlates with the report by Aboussekhra and Al-Sharif (40) that GGR increases in the nontranscribed strand of the GAL10 gene in RAD26Δ cells. Therefore, it is possible that the high rate of TCR in the RPB2 gene reduces GGR in the same locus. This suppression of GGR may reflect either competition for NER factors, or physical hindrance caused by the transcription and/or repair machinery recruited to the transcribed strand.
The fact that TCR plays a primary role in CPD removal at the RPB2 locus is supported by the result that repair drops significantly in H4 R45C cells depleted of RAD26 (Figure 6B). As RAD26 deletion had little effect on NER of the HML locus in both wt and H4 R45C cells (Figure 6C), this result indicates that TCR has little (or no) role in CPD removal from a transcriptionally silent locus, like HML. Thus, the increased NER observed seems to be an effect of the H4 R45 mutation on HML chromatin structure. Indeed, nucleosomes in the HML locus of some Sin mutants are associated with less Sir proteins compared to wt cells (21). Similarly, in the transcriptionally repressed GAL10 locus of H4 R45 mutants, where TCR plays no significant role in repair, the increased NER must reflect a property of the mutant nucleosomes at this locus, which facilitates more rapid repair.
In support of the above notion, genome-wide expression analysis indicates that the enhanced repair observed in H4 R45 mutant cells is not due to increased expression of NER genes. Among the 475 genes that are transcriptionally upregulated in H4 R45H cells, only one NER gene, RAD2, was found to have a >2-fold increase in expression. Interestingly, the microarray data indicate that many of the genes upregulated in H4 R45H cells function in the oxidative or osmotic stress response pathway (unpublished data). Thus, all our results indicate that the increased NER efficiency we observe in the different chromatin loci is a direct effect of altered chromatin structure in the H4 R45 mutants.
In summary, we have shown that single-point mutations in histone H4 (R45C, R45H) of yeast results in increased UV survival, and enhanced NER in specific chromatin loci. The increased NER observed in these mutants primarily reflects an altered chromatin structure, which provides a better substrate for NER complexes than wt cells. Among the other Sin mutants we tested for UV sensitivity, H4 V43I is known to have a less pronounced effect on chromatin structure (17), which may explain the decreased UV resistance compared to H4 R45 mutants. Furthermore, it will be interesting to examine the chromatin structure of H3 T118I mutants, which exhibit a severe Sin phenotype similar to H4 R45 mutants (14,15), but show decreased UV resistance compared to H4 R45 mutants. In conclusion, our results present the first report of enhanced NER in chromatin resulting from a single amino acid change in a core histone and demonstrate how subtle changes in the highly conserved histone sequences can have a major impact on vital cellular functions like DNA repair.
ACKNOWLEDGEMENTS
We are grateful to Dr John Wyrick, Washington State University, for the WY121 yeast strain, for microarray analyses and for helpful discussions throughout the project. We also thank Derek Pouchnik for carrying out the microarray experiments at the WSU Bioinformatics Core Facility. This study was made possible by NIH grant ES04106 from the National Institute of Environmental Health Sciences (NIEHS). Its contents are solely the responsibility of authors and do not necessarily represent the official views of the NIEHS, NIH. Funding to pay the Open Access publication charges for this article was provided by NIH grant ES04106.
Conflict of interest statement. None declared.
REFERENCES
- 1.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis, ASM Press, Washington, DC; 2006. [Google Scholar]
- 2.Peterson CL, Cote J. Cellular machineries for chromosomal DNA repair. Genes Dev. 2004;18:602–616. doi: 10.1101/gad.1182704. [DOI] [PubMed] [Google Scholar]
- 3.Sweder KS, Hanawalt PC. Preferential repair of cyclobutane pyrimidine dimers in the transcribed strand of a gene in yeast chromosomes and plasmids is dependent on transcription. Proc. Natl Acad. Sci. USA. 1992;89:10696–10700. doi: 10.1073/pnas.89.22.10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li S, Smerdon MJ. Dissecting transcription-coupled and global genomic repair in the chromatin of yeast GAL1-10 genes. J. Biol. Chem. 2004;279:14418–14426. doi: 10.1074/jbc.M312004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gillet LC, Scharer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 2006;106:253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 6.Belmont AS, Bruce K. Visualization of G1 chromosomes: a folded, twisted, supercoiled chromonema model of interphase chromatid structure. J. Cell. Biol. 1994;127:287–302. doi: 10.1083/jcb.127.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 8.Gong F, Kwon Y, Smerdon MJ. Nucleotide excision repair in chromatin and the right of entry. DNA Repair. 2005;4:884–896. doi: 10.1016/j.dnarep.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Smerdon MJ, Conconi A. Modulation of DNA damage and DNA repair in chromatin. Prog. Nucleic Acid Res. Mol. Biol. 1999;62:227–255. doi: 10.1016/s0079-6603(08)60509-7. [DOI] [PubMed] [Google Scholar]
- 10.Smerdon MJ, Lieberman MW. Nucleosome rearrangement in human chromatin during UV-induced DNA-reapir synthesis. Proc. Natl Acad. Sci. USA. 1978;75:4238–4241. doi: 10.1073/pnas.75.9.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luger K. Structure and dynamic behavior of nucleosomes. Curr. Opin. Genet. Dev. 2003;13:127–135. doi: 10.1016/s0959-437x(03)00026-1. [DOI] [PubMed] [Google Scholar]
- 12.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 13.Luger K, Richmond TJ. The histone tails of the nucleosome. Curr. Opin. Genet. Dev. 1998;8:140–146. doi: 10.1016/s0959-437x(98)80134-2. [DOI] [PubMed] [Google Scholar]
- 14.Muthurajan UM, Bao Y, Forsberg LJ, Edayathumangalam RS, Dyer PN, White CL, Luger K. Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions. EMBO J. 2004;23:260–271. doi: 10.1038/sj.emboj.7600046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kruger W, Peterson CL, Sil A, Coburn C, Arents G, Moudrianakis EN, Herskowitz I. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev. 1995;9:2770–2779. doi: 10.1101/gad.9.22.2770. [DOI] [PubMed] [Google Scholar]
- 16.Kurumizaka H, Wolffe AP. Sin mutations of histone H3: influence on nucleosome core structure and function. Mol. Cell. Biol. 1997;17:6953–6969. doi: 10.1128/mcb.17.12.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wechser MA, Kladde MP, Alfieri JA, Peterson CL. Effects of Sin- versions of histone H4 on yeast chromatin structure and function. EMBO J. 1997;16:2086–2095. doi: 10.1093/emboj/16.8.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horn PJ, Crowley KA, Carruthers LM, Hansen JC, Peterson CL. The SIN domain of the histone octamer is essential for intramolecular folding of nucleosomal arrays. Nat. Struct. Biol. 2002;9:167–171. doi: 10.1038/nsb762. [DOI] [PubMed] [Google Scholar]
- 19.Flaus A, Rencurel C, Ferreira H, Wiechens N, Owen-Hughes T. Sin mutations alter inherent nucleosome mobility. EMBO J. 2004;23:343–353. doi: 10.1038/sj.emboj.7600047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin AM, Pouchnik DJ, Walker JL, Wyrick JJ. Redundant roles for histone H3 N-terminal lysine residues in subtelomeric gene repression in Saccharomyces cerevisiae. Genetics. 2004;167:1123–1132. doi: 10.1534/genetics.104.026674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fry CJ, Norris A, Cosgrove M, Boeke JD, Peterson CL. The LRS and SIN domains: two structurally equivalent but functionally distinct nucleosomal surfaces required for transcriptional silencing. Mol. Cell. Biol. 2006;26:9045–9059. doi: 10.1128/MCB.00248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li S, Smerdon MJ. Rpb4 and Rpb9 mediate subpathways of transcription-coupled DNA repair in Saccharomyces cerevisiae. EMBO J. 2002;21:5921–5929. doi: 10.1093/emboj/cdf589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 24.Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 25.Kent NA, Bird LE, Mellor J. Chromatin analysis in yeast using NP-40 permeabilised sphaeroplasts. Nucleic Acids Res. 1993;21:4653–4654. doi: 10.1093/nar/21.19.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kent NA, Mellor J. Chromatin structure snap-shots: rapid nuclease digestion of chromatin in yeast. Nucleic Acids Res. 1995;23:3786–3787. doi: 10.1093/nar/23.18.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiss K, Simpson RT. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating type locus HMLalpha. Mol. Cell. Biol. 1998;18:5392–5403. doi: 10.1128/mcb.18.9.5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 29.Li S, Smerdon MJ. Nucleosome structure and repair of N-methylpurines in the GAL1-10 genes of Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:44651–44659. doi: 10.1074/jbc.M206623200. [DOI] [PubMed] [Google Scholar]
- 30.Livingstone-Zatchej M, Marcionelli R, Moller K, de Pril R, Thoma F. Repair of UV lesions in silenced chromatin provides in vivo evidence for a compact chromatin structure. J. Biol. Chem. 2003;278:37471–37479. doi: 10.1074/jbc.M306335200. [DOI] [PubMed] [Google Scholar]
- 31.Smerdon –MJ. DNA repair and the role of chromatin structure. Curr. Opin. Cell. Biol. 1991;3:422–428. doi: 10.1016/0955-0674(91)90069-b. [DOI] [PubMed] [Google Scholar]
- 32.van Gool AJ, Citterio E, Rademakers S, van Os R, Vermeulen W, Constantinou A, Egly JM, Bootsma D, Hoeijmakers H. The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. EMBO J. 1997;16:5955–5965. doi: 10.1093/emboj/16.19.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smerdon MJ, Thoma F. Site-specific DNA repair at the nucleosome level in a yeast minichromosome. Cell. 1990;61:675–684. doi: 10.1016/0092-8674(90)90479-x. [DOI] [PubMed] [Google Scholar]
- 34.Wellinger RE, Thoma F. Nucleosome structure and positioning modulate nucleotide excision repair in the non-transcribed strand of an active gene. EMBO J. 1997;16:5046–5056. doi: 10.1093/emboj/16.16.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smerdon MJ. DNA repair mechanisms and their biological implications. 1989. In Mammalian Cells, Lambert,M.W. and Laval,J. (eds), Plenum Publishing Corp., NewYork, pp. 271–294. [Google Scholar]
- 36.Green CM, Almouzni G. When repair meets chromatin. First in series on chromatin dynamics. EMBO Rep. 2002;3:28–33. doi: 10.1093/embo-reports/kvf005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gong F, Fahy D, Smerdon MJ. Rad4-Rad23 interaction with SWI/SNF links ATP-dependent chromatin remodeling with nucleotide excision repair. Nat. Struct. Mol. Biol. 2006;13:902–907. doi: 10.1038/nsmb1152. [DOI] [PubMed] [Google Scholar]
- 38.Bucceri A, Kapitza K, Thoma F. Rapid accessibility of nucleosomal DNA in yeast on a second time scale. EMBO J. 2006;25:3123–3132. doi: 10.1038/sj.emboj.7601196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu Y, Teng Y, Liu H, Reed SH, Waters R. UV irradiation stimulates histone acetylation and chromatin remodeling at a repressed yeast locus. Proc. Natl Acad. Sci. USA. 2005;102:8650–8655. doi: 10.1073/pnas.0501458102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aboussekhra A, Al-Sharif IS. Homologous recombination is involved in transcription-coupled repair of UV damage in Saccharomyces cerevisiae. EMBO J. 2005;24:1999–2010. doi: 10.1038/sj.emboj.7600665. [DOI] [PMC free article] [PubMed] [Google Scholar]