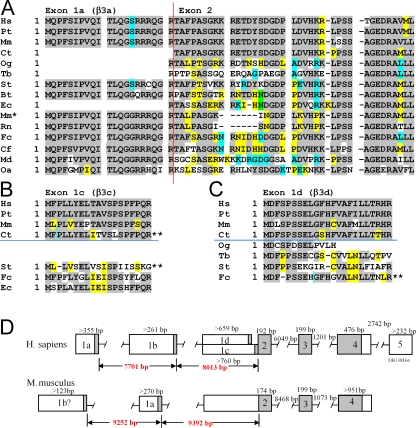
Figure 1.
KCNMB3 N-terminal variation. (A) Predicted β3a (exon 1a) and the initial segment of exon 2 were determined for a range of vertebrate species from searches of publicly available databases, as described in the Materials and methods. Sequences are from the top: human (Hs: Homo sapiens), chimpanzee (Pt: Pan troglodytes), rhesus monkey (Mm: Mulatta macaca), marmoset (Ct: Callithrix jacchus), bushbaby (Og: Otolemur garnettii), treeshrew (Tb: Tupaia belangeri), thirteen-lined squirrel (St: Spermophilus tridecemlineatus), cow (Bt: Bos taurus), horse (Ec: Equus caballus), mouse (Mm': Mus musculus), rat (Rn: Rattus norvegicus), cat (Fc: Felix catus), dog (Cf: Canus lupis familiaris), opossum (Md: Monodelphis domestica), and duckbilled platypus (Oa: Ornithorhynchus anatinus). Exon 1a when attached to exon 2 encodes the β3a isoform. The β3b isoform is generated when exon 1b (not depicted), corresponding to a single methionine, is attached to Exon 2. Vertical red line indicates junction between N-terminal exons and exon 2. Gray highlights residues that are identical among most species. Unshaded residues are unique, while yellow, blue, or green indicate residues shared among a smaller subset of species. (B) Potential N termini with homology to human β3c are aligned. Horizontal blue line separates primate (above) and nonprimate (below) species. The marmoset (Ct) open reading frame (ORF) requires a change in reading frame (indicated by **), while the cat (St) ORF is in a different reading from exon 2. (C) Potential N termini with homology to human β3d are aligned with a horizontal blue line separating primates from other mammals. The cat putative β3d N terminus contained frame shifts and stop codons in the reading frame (**). (D) KCNMB3 gene maps for human and mouse are compared. In contrast to human, the putative mouse 1b coding exon (1b?) that was identified by cloning is positioned upstream of the conserved 1a exon.