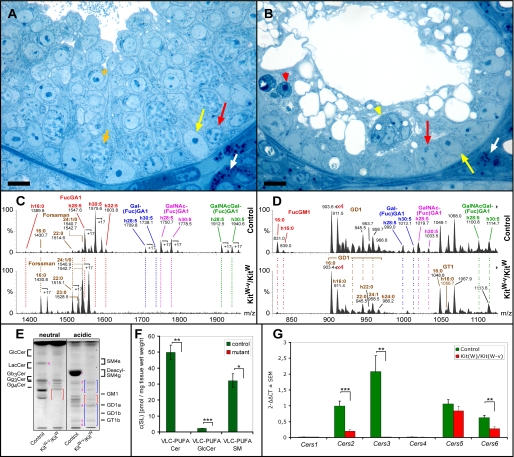
FIGURE 3.
Histological (A and B), sphingolipid (C-F), and Cers mRNA (G) analysis of testes from 4-week-old control (KitW) and KitW-v/KitW mice. A and B, as compared with controls (A), mutant seminiferous tubules (B) are atrophic and contain neither intact spermatocytes nor spermatids. A few functional type A spermatogonia (red arrows) can be detected along the basement membrane. Note, in mutant mice differentiating spermatogonia undergo apoptosis (red arrowhead) as well as Sertoli cells forming multinuclear aggregates (yellow arrowhead). Semithin Epon sections were stained with methylene blue-Azur II. White arrows, Leydig cells; yellow arrows, Sertoli cell nuclei; short orange arrow, adluminal primary pachytene spermatocyte; orange asterisk, round spermatid; bar, 10 μm. C, and D, nano-electrospray ionization-tandem mass spectrometry characterization of neutral (C) and acidic (D) complex GSLs from control and KitW-v/KitW (mutant) testis. Complex neutral GSLs were detected with a precursor ion scan of m/z +204. Signals for fucosylated VLC-PUFA GSLs (FucGA1, Gal(Fuc)GA1, GalNAc(Fuc)GA1, and GalNAc-Gal(Fuc)GA1) dominate in control sample with signals of Forssman lipid not exceeding 12% of relative intensity. In mutant (KitW-v/KitW) testis, signals for fucosylated VLC-PUFA GSLs are lacking. Signals for Forssman lipid are present and become base peak. Complex gangliosides were detected with a precursor ion scan of m/z -87. Signals for fucosylated VLC-PUFA gangliosides (Gal(Fuc)GM1, GalNAc(Fuc)GM1, and GalNAcGal(Fuc)GM1) are not present in mutant sample, whereas signals for FucGM1(d18:1,16:0), GD1, and GT1 are detected. E, TLC of testicular GSLs from control (KitW) and KitW-v/KitW testes. GSLs were split into a neutral and an acidic fraction and stained after separation with orcinol. Lanes correspond to 20 mg of tissue wet weight. Red and blue brackets denote the migration areas of polyenoic fucosylated GSLs and nonpolyenoic “classical” gangliosides, respectively. Note, bands for neutral polyenoic fucosylated GSLs are not detectable in KitW-v/KitW testes and corresponding bands for acidic polyenoic GSLs are reduced. Bands for Forssman lipid (band 1) and nonpolyenoic classical gangliosides GM3 (band 2), GM2 (band 3), GD1a (band 4), GT1b (band 5), and GQ1b (band 6) appear not to be altered. Seminolipid (SM4g) (band 7) and its precursor 1-alkyl-2-acyl-3-O-β-d-galactosyl-sn-glycerol (GalEAG) (band 8), detected here as deacyl compounds, are also missing in KitW-v/KitW testes. F, mass spectrometric quantification of the VLC-PUFA sphingolipids ceramide, GlcCer, and sphingomyelin from control (KitW) and KitW-v/KitW testes. Internal standards for Cer, GlcCer, and SM were added to lipid extracts. Cer and GlcCer were detected with the precursor ion mode m/z +264 selective for d18:1 and t18:0 sphingoid base containing sphingolipids. SM was detected with the precursor ion mode m/z +184 selective for the phosphorylcholine head group. G, quantitative real time-PCR of Cers mRNAs from control (KitW) and KitW-v/KitW testes. The mRNA was isolated from 4-week-old control (KitW) and KitW-v/KitW testes, transcribed into cDNA, and subjected to qRT-PCR. ΔCT values (CT(Cers)-CT(GAPDH)) obtained from qRT-PCR were normalized to ΔCT of control Cers5 mRNA (ΔΔCT). F and G, Student's t test, *, p < 0.05; **, p < 0.01; ***, p < 0.001.