Abstract
Subunit a plays a key role in promoting H+ transport and the coupled rotary motion of the subunit c ring in F1F0-ATP synthase. H+ binding and release occur at Asp-61 in the middle of the second transmembrane helix (TMH) of F0 subunit c. H+ are thought to reach Asp-61 via aqueous pathways mapping to the surfaces of TMHs 2–5 of subunit a based upon the chemical reactivity of Cys substituted into these helices. Here we substituted Cys into loops connecting TMHs 1 and 2 (loop 1–2) and TMHs 3 and 4 (loop 3–4). A large segment of loop 3–4 extending from loop residue 192 loop to residue 203 in TMH4 at the lipid bilayer surface proved to be very sensitive to inhibition by Ag+. Cys-161 and -165 at the other end of the loop bordering TMH3 were also sensitive to inhibition by Ag+. Further Cys substitutions in residues 86 and 93 in the middle of the 1–2 loop proved to be Ag+-sensitive. We next asked whether the regions of Ag+-sensitive residues clustered together near the surface of the membrane by combining Cys substitutions from two domains and testing for cross-linking. Cys-161 and -165 in loop 3–4 were found to cross-link with Cys-202, -203, or -205, which extend into TMH4 from the cytoplasm. Further Cys at residues 86 and 93 in loop 1–2 were found to cross-link with Cys-195 in loop 3–4. We conclude that the Ag+-sensitive regions of loops 1–2 and 3–4 may pack in a single domain that packs at the ends of TMHs 3 and 4. We suggest that the Ag+-sensitive domain may be involved in gating H+ release at the cytoplasmic side of the aqueous access channel extending through F0.
The H+-transporting F1F0-ATP synthases of oxidative phosphorylation utilize the energy of a transmembrane electrochemical gradient of H+ or Na+ to mechanically drive the synthesis of ATP via two coupled rotary motors in the F1 and F0 sectors of the enzyme (1–3). In the intact enzyme, ATP synthesis or hydrolysis takes place in the F1 sector at the surface of the membrane with synthesis coupled to H+ transport through the transmembrane F0 sector. Homologous enzymes are found in mitochondria, chloroplasts, and many bacteria (4). In Escherichia coli and other eubacteria, F1 consists of five subunits in an α3β3γ1δ1ε1 stoichiometry (4). F0 is composed of three subunits in a likely ratio of a1b2c10 in E. coli and Bacillus subtilis PS3 or a1b2c11 in the Na+-translocating Ilyobacter tartaricus ATP synthase (3, 5–7) and may contain as many as 15 c subunits in other bacterial species (8). Subunit c spans the membrane as a helical hairpin with the first TMH2 on the inside and the second TMH on the outside of the c ring (7, 9, 10). A high resolution x-ray structure of the I. tartaricus c11 ring has revealed the sodium binding site at the periphery of the ring with chelating groups to the Na+ extending from two interacting subunits (7). The essential I. tartaricus Glu-65 in the Na+-chelating site corresponds to E. coli Asp-61. In the H+-transporting E. coli enzyme, Asp-61 at the center of the second TMH is thought to undergo protonation and deprotonation as each subunit of the c ring moves past a stationary subunit a. In the complete membranous enzyme, the rotation of the c ring is proposed to be driven by H+ transport at the subunit a/c interface with ring movement then driving rotation of subunit γ within the α3β3 hexamer of F1 to cause conformational changes in the catalytic sites leading to synthesis and release of ATP (1–3).
Subunit a folds in the membrane with five TMHs and is thought to provide access channels to the proton-binding Asp-61 residue in the c ring (11–14). Interaction of the conserved Arg-210 residue in aTMH4 with cTMH2 is thought to be critical during the deprotonation-protonation cycle of cAsp-61 (15–17), and aTMH4 and cTMH2 are known to pack in parallel to each other based upon cross-linking (18). Little is known about the structure or three-dimensional arrangement of the TMHs in subunit a. Based initially upon the position of several sets of second site suppressor mutations, we recently introduced pairs of Cys into putatively apposing TMHs and tested for zero-length cross-linking with disulfide bond formation (19). Cross-links were found with eight different Cys pairs and define a juxtaposition of TMHs 2–3, 2–4, 2–5, 3–4, 3–5, and 4–5 packing in a four-helix bundle (19).
The differential reactivity of cysteines introduced by sitedirected mutagenesis has been used as a means of probing aqueous accessible regions of several membrane proteins. The reagents used, including methanethiosulfonate (MTS) derivatives (20, 21), NEM (22, 23), and Ag+ (24, 25), react preferentially with the ionized, thiolate form of the Cys side chain and thus can serve as an indicator of the polarity of the environment. Previously we probed Cys residues introduced into the five TMHs of subunit a for aqueous accessibility based upon their reactivity with NEM and Ag+ (26–28). Two regions of aqueous access were found with distinctly different properties. One region extending from Asn-214 and Arg-210 near the center of the membrane to the cytoplasmic surface of TMH4 contains Cys residues that are sensitive to inhibition by both NEM and Ag+. A second set of Ag+-sensitive but NEM-inaccessible residues mapped to the opposite face and periplasmic side of TMH4. Ag+-sensitive but NEM-insensitive residues extending from the center of the membrane to the periplasmic surface were also found in TMHs 2, 3, and 5. The Ag+-sensitive residues in TMHs 2, 3, 4, and 5 cluster at the interior of the fourhelix bundle predicted by cross-linking (19) and could interact to form a continuous aqueous pathway extending from the periplasmic surface to the center of the membrane. In the experiments reported here, we probed the properties of Cys residues introduced at the cytoplasmic boundary of TMH4 and were surprised to discover an extended cytoplasmic region that was sensitive to Ag+ inhibition. We ultimately introduced Cys substitutions throughout the entire 3–4 loop and then extended mapping to the 1–2 loop. Ag+-sensitive Cys residues were found at both ends of the 3–4 loop and in the middle of the 1–2 loop. We next asked whether the domains of Ag+-sensitive residues clustered together and packed near the surface of the membrane by combining Cys substitutions from two domains and testing for cross-linking. The Ag+-sensitive regions of loops 1–2 and 3–4 proved to be cross-linkable via Cys introduced near the cytoplasmic lipid bilayer interface at the ends of TMHs 3 and 4. We suggest that this Ag+-sensitive domain(s) may be involved in gating H+ release to the cytoplasm from the aqueous access channel extending through F0.
EXPERIMENTAL PROCEDURES
Construction of Cys-substituted Mutants—The cysteine substitutions generated here were transferred into plasmid pCMA113 (26), which contains a hexahistidine tag on the C terminus of subunit a and structural genes encoding F1F0 subunits from which all endogenous Cys had been substituted by Ala or Ser (29). Cys substitutions were introduced by a two-step PCR method using a synthetic oligonucleotide, which contained the codon change, and two wild type primers (30). PCR products were transferred into pCMA113 directly using unique HindIII (870) or PflMI (1136) and BsrGI (1913) sites (see Ref. 31 for nucleotide numbering). All mutations were confirmed by sequencing the cloned fragment through the ligation junctions. All experiments were performed with the Cys-substituted derivatives of plasmid pCMA113 transformed into the unc (atp) operon deletion host strain JWP292 (5). All plasmid transformant strains were tested for growth on succinate and glucose as described previously (26).
Membrane Preparation—Plasmid transformants of strain JWP292 were grown in M63 minimal medium containing 0.6% glucose, 2 mg/liter thiamine, 0.2 mm uracil, 1 mm l-arginine, 0.02 mm dihydroxybenzoic acid, and 0.1 mg/ml ampicillin supplemented with 10% LB medium and harvested in the late exponential phase of growth (5). Cells were suspended in TMG-acetate buffer (50 mm Tris acetate, 5 mm magnesium acetate, 10% glycerol, pH 7.5) containing 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and 0.1 mg/ml DNase I and disrupted by passage through a French press at 1.38 × 108 newtons/m2, and membranes were prepared as described previously (32). The final membrane preparation was suspended in TMG-acetate buffer and stored at –80 °C. Protein concentrations were determined using a modified Lowry assay (33).
ATP-driven Quenching of ACMA Fluorescence—Membranes were suspended in 3.2 ml of HMK-nitrate buffer (10 mm Hepes-KOH, 1 mm Mg(NO3)2, 10 mm KNO3, pH 7.5). ACMA was added to 0.3 μg/ml final concentration, and 30 μl of 0.1 m ATP, pH 7.0, was added to initiate quenching of fluorescence. The reaction was terminated by addition of 8 μl of 288 μm nigericin (0.5 μg/ml final concentration). The level of fluorescence obtained after addition of nigericin was normalized to 100% in calculating the percent quenching due to ATP-driven proton pumping. For AgNO3 treatment, 160 μl of membranes at 10 mg/ml were suspended in HMK-nitrate buffer containing 40 μm AgNO3 and incubated at room temperature for 15 min before carrying out the quenching assay. For NEM treatment, membranes at 10 mg/ml in TMG-acetate buffer were treated with 5 mm NEM for 15 min at room temperature and then diluted into HMK-nitrate buffer before carrying out the same quenching assay.
Cross-linking with Bis-MTS Reagents—Cys-substituted membranes were diluted to 10 mg/ml in TMG-acetate buffer at pH 8.5. The bis-MTS reagents M2M and M4M (Toronto Research Chemicals) were dissolved in dimethyl sulfoxide and added to the membranes to a final concentration of 250 μm. The reaction was incubated for 10 min at room temperature and then treated with 5 μl of 0.5 m Na2EDTA, pH 8.0, and incubated an additional 15 min at room temperature. To reduce and reverse cross-link formation, β-mercaptoethanol was added to a final concentration of 4% by volume, or alternatively dithiothreitol was added to a concentration of 20 mm, and the sample was incubated at room temperature for 15 min prior to dilution into an equal volume of 2× SDS sample buffer (0.125 m Tris, pH 6.8, 4% SDS, 20% (v/v) glycerol) and then incubated at room temperature for 1 h before analysis by SDS-PAGE. Modification by 1 mm DABMI (Molecular Probes Inc.) was also carried out in SDS sample buffer for 1 h prior to SDS-PAGE.
SDS Electrophoresis and Immunoblotting—Samples were run using the SDS-PAGE system described by Laemmli (34). The separating gel contained 15% acrylamide and 0.71% bisacrylamide in 0.375 m Tris at pH 8.8 with 0.125% SDS. The stacking gel was 3% acrylamide and 0.9% bisacrylamide in 0.125 m Tris at pH 6.8 with 0.1% SDS. The running buffer was 0.025 m Tris, 0.192 m glycine, 0.2% SDS adjusted to pH 8.3. The gel was run for 2 h at 75 V through the stacking gel and then for 25 h at a constant 15 mA. Proteins in the separatory gel were transferred to polyvinylidene difluoride (PVDF) membrane by applying 75 V for 1.5 h using 0.192 m glycine, 0.025 m Tris, 20% MeOH buffer adjusted to pH 8.3 (35). Western blotting was performed after incubation of the PVDF membrane with 5 mg/ml of GE Healthcare Blocking Agent in 1× PBS-Tween (137 mm NaCl, 1.5 mm NaH2PO4, 6.5 mm Na2HPO4, 0.1% Tween 20) for 1 h at room temperature or alternatively overnight at 4 °C. Rabbit antiserum directed against the first 10 amino acids of subunit a was preabsorbed to E. coli membranes lacking F0 to reduce immunoartifacts (36). The primary antiserum against subunit a was diluted 1:5000 in 1× PBS-Tween with 2% bovine serum albumin and incubated with the PVDF membrane for 1 h at room temperature. Anti-rabbit horseradish peroxidase-linked secondary antibody was diluted 1:40,000 in 1× PBS-Tween and incubated with the PVDF membrane for 1 h at room temperature. The PVDF membrane was incubated for 4.5 min with equal volumes of Super Signal West Pico Chemiluminescent Substrates (Pierce). The PVDF membrane was then exposed to film to visualize the protein.
RESULTS
Properties of Cys Substitutions in Loops 1–2 and 3–4 of Subunit a—In this study, Cys substitutions in the 1–2 and 3–4 loops of subunit a were generated in a plasmid encoding subunits of F1 and F0 in which the endogenous Cys residues were substituted by Ala or Ser (26). These plasmids were transformed into strain JWP292, which carries a chromosomal deletion of the entire unc (atp) operon. The growth of these strains on minimal medium was compared using glucose or succinate as a carbon sources (Table 1). The Cys substitutions reported here had minimal effects on growth with either carbon source. Additionally ATP-driven quenching of ACMA fluorescence was performed on inside-out membrane vesicles of the substituted strains to evaluate ATPase-coupled H+ pumping function (Table 1). The quenching response for most mutants was close to that observed with the control membranes, i.e. maximal quenching in the range of 70–80%. Six substitutions at positions 171, 174, 177, 182, 192, and 195 did lead to significant reductions in quenching into a range of 40–60%, but each of these grew nearly as well as wild type via oxidative phosphorylation on succinate minimal medium.
TABLE 1.
Functional properties of subunit a substitutions in the 1-2 and 3-4 loops
The properties of substitutions at residues 62, 96-105, 159-164, and 200-203 were also reported elsewhere (28).
| Location and mutation | Growth on glucosea | Growth on succinateb | Quenching with ATPc |
|---|---|---|---|
| % | |||
| 1-2 loop | |||
| S62C | 102 | 2.5 | 78 ± 1 |
| V63C | 101 | 2.0 | 85 ± 1 |
| A64C | 100 | 2.2 | 82 ± 0 |
| K65C | 107 | 2.2 | 80 ± 2 |
| K66C | 105 | 2.2 | 71 ± 2 |
| A67C | 100 | 2.0 | 77 ± 1 |
| T68C | 104 | 2.0 | 80 ± 0 |
| S69C | 100 | 2.0 | 73 ± 4 |
| G70C | 101 | 2.1 | 67 ± 3 |
| V71C | 99 | 2.1 | 71 ± 4 |
| P72C | 101 | 2.0 | 70 ± 6 |
| G73C | 108 | 2.0 | 73 ± 3 |
| K74C | 100 | 2.2 | 84 ± 0 |
| F75C | 105 | 2.0 | 70 ± 7 |
| Q76C | 107 | 2.0 | 84 ± 1 |
| T77C | 103 | 2.0 | 69 ± 7 |
| A78C | 103 | 2.0 | 70 ± 5 |
| I79C | 101 | 2.0 | 68 ± 4 |
| E80C | 99 | 2.0 | 84 ± 1 |
| L81C | 111 | 2.0 | 75 ± 4 |
| V82C | 99 | 1.9 | 70 ± 5 |
| I83C | 108 | 2.0 | 75 ± 3 |
| G84C | 102 | 1.8 | 66 ± 3 |
| F85C | 108 | 2.0 | 74 ± 1 |
| V86C | 102 | 2.0 | 68 ± 2 |
| N87C | 112 | 2.0 | 76 ± 1 |
| G88C | 104 | 2.0 | 71 ± 1 |
| S89C | 99 | 2.3 | 71 ± 2 |
| V90C | 102 | 2.3 | 68 ± 5 |
| K91C | 98 | 2.0 | 71 ± 2 |
| D92C | 105 | 2.1 | 66 ± 4 |
| M93C | 105 | 2.0 | 67 ± 3 |
| Y94C | 100 | 2.1 | 70 ± 3 |
| H95C | 103 | 2.2 | 67 ± 6 |
| G96C | 112 | 2.6 | 77 ± 1 |
| K97C | 100 | 2.1 | 78 ± 3 |
| S98C | 100 | 2.2 | 76 ± 2 |
| K99C | 111 | 2.4 | 76 ± 2 |
| L100C | 100 | 2.2 | 73 ± 3 |
| I101C | 100 | 2.4 | 76 ± 1 |
| A102C | 105 | 2.4 | 74 ± 2 |
| 3-4 loop | |||
| I159C | 101 | 2.2 | 69 ± 4 |
| L160C | 100 | 2.3 | 64 ± 2 |
| I161C | 99 | 2.3 | 70 ± 4 |
| L162C | 99 | 2.2 | 67 ± 2 |
| F163C | 99 | 2.2 | 69 ± 8 |
| Y164C | 88 | 2.0 | 74 ± 4 |
| S165C | 99 | 2.1 | 69 ± 7 |
| I166C | 100 | 2.2 | 73 ± 0 |
| K167C | 99 | 2.1 | 74 ± 1 |
| M168C | 99 | 2.4 | 70 ± 5 |
| K169C | 99 | 2.3 | 70 ± 5 |
| G170C | 99 | 2.5 | 73 ± 3 |
| I171C | 100 | 2.6 | 47 ± 6 |
| G172C | 100 | 2.1 | 69 ± 4 |
| G173C | 101 | 2.6 | 69 ± 3 |
| F174C | 101 | 2.4 | 55 ± 16 |
| T175C | 100 | 2.3 | 73 ± 5 |
| K176C | 100 | 2.3 | 70 ± 1 |
| E177C | 100 | 2.1 | 58 ± 14 |
| L178C | 99 | 2.5 | 68 ± 3 |
| T179C | 99 | 2.4 | 70 ± 0 |
| L180C | 100 | 2.3 | 75 ± 4 |
| Q181C | 100 | 2.1 | 73 ± 0 |
| P182C | 98 | 2.2 | 59 ± 5 |
| F183C | 99 | 2.2 | 74 ± 1 |
| N184C | 99 | 2.4 | 75 ± 0 |
| H185C | 104 | 2.3 | 68 ± 1 |
| W186C | 100 | 2.7 | 65 ± 3 |
| A187C | 101 | 2.7 | 65 ± 3 |
| F188C | 100 | 2.7 | 64 ± 0 |
| I189C | 101 | 2.3 | 64 ± 0 |
| P190C | 101 | 2.5 | 69 ± 4 |
| V191C | 106 | 2.5 | 63 ± 5 |
| N192C | 99 | 1.9 | 42 ± 3 |
| L193C | 91 | 2.2 | 70 ± 8 |
| I194C | 95 | 2.2 | 67 ± 5 |
| L195C | 94 | 2.1 | 59 ± 3 |
| E196C | 89 | 2.0 | 73 ± 8 |
| G197C | 97 | 2.2 | 75 ± 6 |
| V198C | 92 | 2.2 | 74 ± 5 |
| S199C | 99 | 2.3 | 71 ± 3 |
| L200C | 99 | 2.2 | 72 ± 3 |
| L201C | 99 | 2.0 | 73 ± 1 |
| S202C | 97 | 2.2 | 69 ± 1 |
| K203C | 97 | 2.1 | 67 ± 2 |
Yield in liquid minimal medium containing 0.04% glucose expressed as a percentage of growth relative to the cysteine-less control.
Colony size after incubation for 72 h on minimal medium plates containing 22 mm succinate. Colonies from the cysteine-less control strain show average colony sizes of 2.4 ± 0.3 mm.
Measured in a buffer containing 10 mm Hepes-KOH, pH 7.5, 1 mm Mg(NO3)2, 10 mm KNO3.
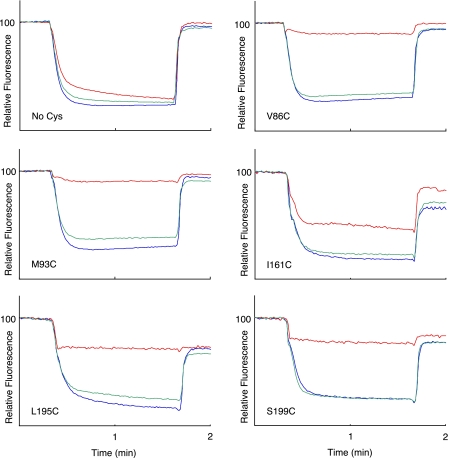
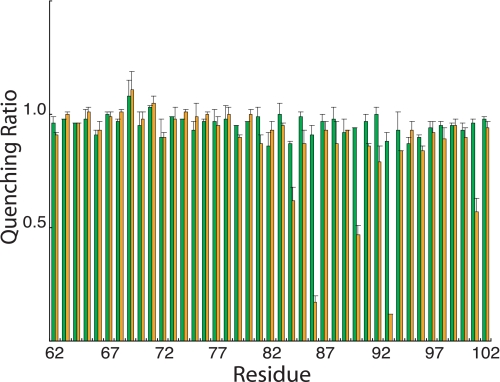
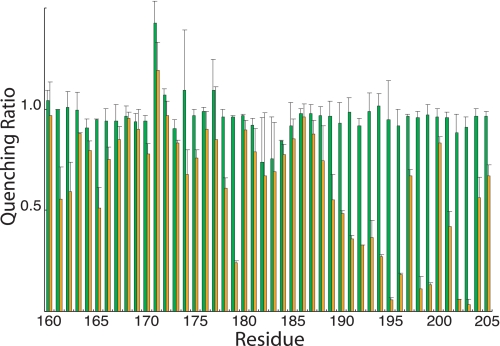
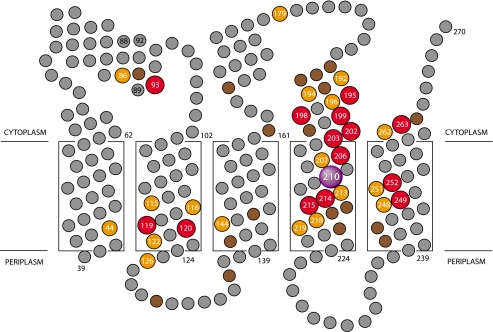
Ag+ Inhibition of ATPase-coupled H+ Transport in Cys-substituted 1–2 and 3–4 Loops—Inside-out membrane vesicles with Cys substitutions in the 1–2 or 3–4 loops were treated with 40 μm AgNO3 or 5 mm NEM in chloride-free assay buffer, and the effects on ATPase-coupled H+ transport were measured by the quenching of ACMA fluorescence. Examples for five Ag+-sensitive but NEM-insensitive mutants are shown in Fig. 1. In four of the five mutants shown, Ag+ caused an approximate 90% reduction in the quenching response. In the case of the I161C substitution in loop 3–4, Ag+ reduced the quenching response by ∼50%. In contrast to the observed Ag+ inhibition, NEM had negligible inhibitory effects on all substitutions examined in this study. The complete surveys of the Ag+-sensitivity and NEM-sensitivity of cysteine substituted into loops 1–2 and 3–4 are shown in Figs. 2 and 3, respectively. The positions of the Ag+-sensitive residues in a two-dimensional topological map of subunit a are shown in Fig. 4. The most striking distribution of multiple Ag+-sensitive residues maps to the C-terminal end of loop 3–4 and extends into the cytoplasmic end of TMH4 (Figs. 3 and 4). A single Ag+-sensitive Cys was found in the middle of loop 3–4. In addition, three cysteines showing moderate sensitivity to Ag+ (45–50% inhibition) were found at the N-terminal end of loop 3–4. In loop 1–2, Ag+ sensitivity was confined to three positions in the middle of the loop, i.e. residues 86, 90, and 93. The restriction of Ag+-sensitive Cys to a few localized areas of the loops led us to ask whether the regions interacted physically in the folded structure of the protein and to the crosslinking experiments described below.
FIGURE 1.
Differing sensitivity of Cys substitutions in 1–2 loop and 3–4 loop to Ag+ and NEM inhibition. A 160-μl aliquot of membranes at 10 mg/ml in TMG-acetate were treated with 5 mm NEM for 15 min at room temperature and then diluted into 3.2 ml of HMK-nitrate buffer containing 0.3 μg/ml ACMA. Alternatively membranes were diluted into HMK-nitrate buffer, and AgNO3 was added to 40 μm for 15 min at room temperature prior to addition of ACMA. ATP was added to 0.94 mm at 20 s, and the uncoupler nigericin was added to 0.5 μg/ml at 100 s. The traces indicate no treatment (blue), NEM treatment (green), or Ag+ treatment (red). The substitution tested is indicated in each panel.
FIGURE 2.
NEM and Ag+ sensitivity of Cys substitutions in the 1–2 loop. Results are presented as the ratios of the quenching response in the presence of Ag+ or NEM to the quenching response in the absence of either reagent. The green bars represent the quenching ratio ±NEM treatment, whereas the orange bars represent the quenching ratio ±Ag+ treatment. Each bar represents the average ratio from n ≥ 2 determinations ±S.D.
FIGURE 3.
NEM and Ag+ sensitivity of Cys substitutions in the 3–4 loop. Results are presented as the ratios of the quenching response in the presence of Ag+ or NEM to the quenching response in the absence of either reagent. The green bars represent the quenching ratio ±NEM treatment, whereas the orange bars represent the quenching ratio ±Ag+ treatment. Each bar represents the average ratio from n ≥ 2 determinations ±S.D.
FIGURE 4.
Position of Ag+-sensitive residues in a two-dimensional topological map of subunit a. The suggested regions of α-helical secondary structure depicted in the 1–2 and 3–4 loops are based upon NMR chemical shift analysis of subunit a in chloroform-methanol-H2O (4:4:1) solvent (53). Residues that are most sensitive to inhibition by Ag+ are shown in red (>85% inhibition), orange (66–85% inhibition), and brown (46–65% inhibition). The relative depth of placement of the five TMHs of subunit a in the lipid bilayer is based upon cross-linking experiments as described elsewhere (19).
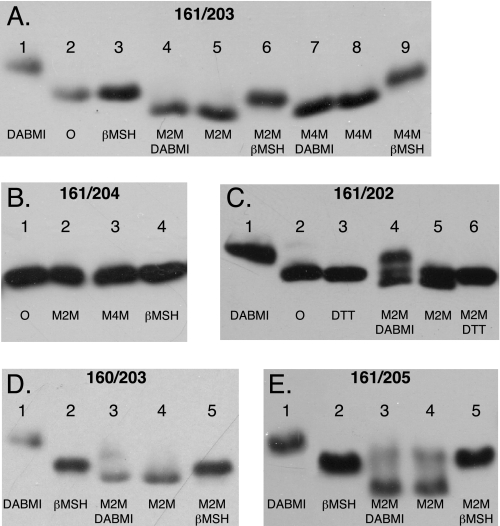
The Ag+-sensitive N- and C-terminal Regions of Loop 3–4 Can Be Cross-linked—To test whether the Ag+-sensitive N- and C-terminal regions of loop 3–4 interacted with each other, we introduced Cys into both regions and tested for cross-linking with the bis-MTS reagents M2M and M4M. These reagents cross-link between two cysteine sulfhydryls with formation of bis-disulfide bonds and insertion of a -S-(CH2)n-S-flexible spacer group where n = 2 for M2M and n = 4 for M4M (21). Cross-link formation was detected by a shift in the mobility of subunit a during SDS-PAGE where the cross-linked product shows greater mobility apparently because of a more compact structure (19). The results of several double Cys combinations with the I161C substitution, which demonstrated the 50% inhibition of ACMA quenching by Ag+ in Fig. 1, are shown in Fig. 5. The general protocol of the experiment is shown in Fig. 5A for the I161C/K203C double Cys substitution. The migration of untreated and β-mercaptoethanol-reduced subunit a is shown in lanes 2 and 3. When the unmodified double Cys substitution is treated with DABMI (lane 1), subunit a migrates to a higher apparent molecular weight due to the addition of DABMI molecules to the two Cys residues (DABMI Mr = 320). Treatment with M2M or M4M causes a shift in the subunit a band to greater mobility (lanes 5 and 8), and the cross-linking prevents subsequent reaction with DABMI in SDS sample buffer (lanes 4 and 7). The M2M and M4M cross-linked products are reduced, and the original band migration is restored following treatment with β-mercaptoethanol (lanes 6 and 9). The specificity of the reaction is demonstrated in the experiment shown in Fig. 5B where neither M2M nor M4M cause cross-linking with the I161C/P204C double Cys substitution. M2M treatment of the I161C/S202C double Cys was concluded to result in partial cross-linking and a smaller shift in mobility of the subunit a band; the shifted band was immune to reaction with DABMI (Fig. 5C). Similar results, indicative of cross-linking, were seen with the L160C/K203C and I161C/V205C double Cys-substituted mutants (Fig. 5, D and E). In these cases M2M appears to also react with a minor fraction of the protein to form monovalent adducts, which increase the apparent molecular weight of the protein, without further reaction of the second thiosulfonate group to form a cross-linked product. A summary of results for the double Cys substitutions tested in these areas of the protein is given in Table 2. Cu2+(phenanthroline)2 failed to catalyze cross-link formation with any of the Cys pairs listed in Table 2 that were positive in cross-link with M2M (data not shown).
FIGURE 5.
Cross-link formation between pairs of Cys substitutions placed in the two ends of the 3–4 loop. Membranes from the double Cys substitutions shown were treated with the reagents shown in each lane in the sequence indicated as described under “Experimental Procedures.” Following SDS-PAGE, Western blots were probed with anti-subunit a antibody. Cross-link formation led to increased electrophoretic mobility and a downward band shift as discussed in the text. A, I161C/K203C membranes; B, I161C/204 membranes; C, I161C/P204C membranes; D, L160C/K203C membranes; E, I161C/V205C membranes. βMSH, β-mercaptoethanol; DTT, dithiothreitol; O, no treatment.
TABLE 2.
Pairs of double Cys substitutions tested for cross-linking between ends of the 3-4 loop
+ indicates cross-link formed after M2M treatment; 0 indicates cross-link not formed after M2M treatment.
|
Residues at N terminus of loop
|
Residues at C terminus of loop
|
||||||
|---|---|---|---|---|---|---|---|
| 202 | 203 | 204 | 205 | 206 | 207 | 208 | |
| 155 | 0 | 0 | 0 | ||||
| 157 | 0 | +a | 0 | ||||
| 158 | 0 | 0 | 0 | 0 | |||
| 159 | 0 | 0 | 0 | 0 | |||
| 160 | 0 | + | 0 | + | |||
| 161 | + | + | 0 | + | |||
| 163 | 0 | 0 | |||||
| 165 | + | 0 | |||||
+ indicates cross-link formed after M4M treatment but not after M2M treatment.
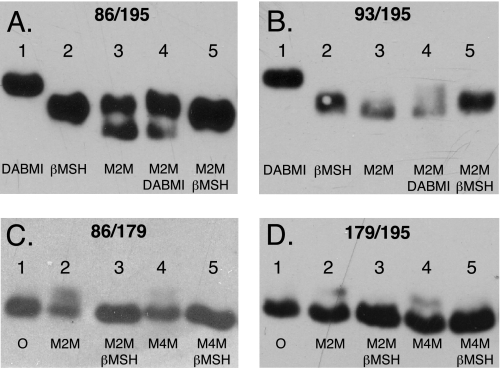
The Ag+-sensitive Central Region of Loop 1–2 Can Be Cross-linked to the C-terminal Ag+-sensitive Region of Loop 3–4—To test whether the Ag+-sensitive regions in the center of loop 1–2 interacted with the C-terminal Ag+-sensitive region of loop 3–4, Cys substitutions were introduced into residues in both regions, and the double Cys substitutions were tested for cross-linking with the M2M and M4M. The positive cross-links seen with the V86C/L195C and M93C/L195C double Cys pairs are shown in Fig. 6. Based upon the intensities of the shifted versus non-shifted bands in the M2M-treated V86C/L195C sample, ∼50% of the subunit a was cross-linked. The two Cys residues in the non-cross-linked V86C/L195C band probably reacted monovalently with M2M. This is indicated by the lack of reaction of both M2M-treated sample bands on subsequent treatment with DABMI (lane 5). We attempted and were unable to cross-link the Ag+-sensitive T179C residue in the middle of loop 3–4 with V86C, M93C (not shown), or L195C. The upward shift seen for a fraction of the V86C/T179C and T179C/L195C M2M-treated samples in Fig. 6 is likely due to monovalent M2M adduct formation with the substituted cysteine(s). We were also unable to cross-link V86C or T179C with Y263C, which is positioned at the cytoplasmic end of TMH5 (not shown). Cu2+(phenanthroline)2 failed to catalyze cross-link formation with any of the Cys pairs listed in Table 2 that were positive in cross-link with M2M (data not shown).
FIGURE 6.
Cross-link formation between Cys substitutions in the center of the 1–2 loop and C-terminal end of the 3–4 loop. Membranes from the double Cys substitutions were treated with the reagents shown in each lane in the sequence indicated as described under “Experimental Procedures.” Following SDS-PAGE, Western blots were probed with anti-subunit a antibody. Cross-link formation led to increased electrophoretic mobility and a downward band shift as discussed in the text. A, V86C/L195C membranes; B, M93C/L195C membranes; C, V86C/T179C membranes; D, T179C/L195C membranes. βMSH, β-mercaptoethanol; O, no treatment.
DISCUSSION
Prior to this study, we reported Ag+-sensitive Cys residues in the five TMHs of subunit a that we concluded were likely to be accessed via an aqueous pathway (26–28). As pointed out by others (24), the ionic radius of Ag+ is close to that of Na+ or H3O+, and hence it may be small enough to permeate waterlined channels. This difference in size may account for its more potent inhibitory properties versus NEM as was documented by direct NEM reactivity assays in previous studies (26, 27). In these previous studies, one set of Ag+- and NEM-sensitive Cys residues was found to cluster on one face on the cytoplasmic side of TMH4 (26) and was proposed to provide an outlet pathway to the cytoplasm (Fig. 4). Other Ag+-sensitive and NEM-insensitive Cys residues tended to cluster toward the periplasmic side of TMHs 2–5 at the center of a proposed four-helix bundle (26–28), and these interacting helices were proposed to provide an aqueous access pathway from the periplasm to the center of the membrane. In this study Ag+-sensitive Cys substitutions were found to extend into localized regions of the 3–4 cytoplasmic loop near the surface of the membrane lipid bilayer. The largest set of Ag+-sensitive residues clustered in a span of 12–15 residues at the cytoplasmic end of TMH4 (Fig. 4). A second smaller set of moderately Ag+-sensitive residues was found at the cytoplasmic boundary extending from TMH3. Finally as the study was expanded, a third set of Ag+-sensitive residues was found at the center of the 1–2 loop.
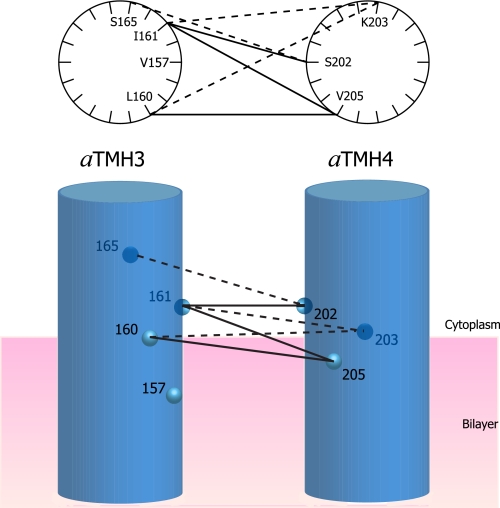
The regions of Ag+-sensitive residues in the 1–2 and 3–4 loops appear to pack within short distances of each other. The packing proximity was indicated by cross-link formation between pairs of Cys introduced at the N- and C-terminal ends of the 3–4 loop and between pairs of Cys introduced in the central region of the 1–2 loop and C-terminal end of the 3–4 loop. The bis-MTS compounds used in these studies would insert short flexible spacers between the cross-linkable Cys pairs, i.e. 5.2 Å for the -S-(CH2)2-S-spacer in M2M (21). Transmembrane helices in other membrane proteins often extend beyond the 30-Å hydrophobic core of the lipid bilayer, and it seems possible that the cross-linkable regions at the ends of the 3–4 loop may be between helical extensions of TMH3 and TMH4 into the head group region of the phospholipid bilayer. The cross-linkable residues are confined to one side of α-helical wheel models for both of these regions (Fig. 7). Cys substituted in TMHs 3 and 4 at positions 148 and 219 at the center of the lipid bilayer can also be cross-linked with Cu2+ (19), suggesting that these TMHs are likely to pack in parallel as they extend through the membrane. Cys substitutions in the N-terminal half of the 3–4 loop can be cross-linked to subunit c via a bifunctional photoactivable maleimide cross-linker (37), supporting the proximal positioning of this loop region to the surface of the membrane.
FIGURE 7.
Cross-link formation between modeled α-helical faces of aTMH3 and aTMH4 as they emerge from the cytoplasmic face of the membrane. The double Cys pairs that cross-link with M2M are tabulated in Table 2 and here shown connected by lines. The light or dark circles are shown to distinguish the front and back faces of the cylinders. The solid versus dashed lines are intended to distinguish cross-linking to the front and back faces of the α-helical wheels or cylinders. The M4M cross-link between 157 and 203 is not connected by a line.
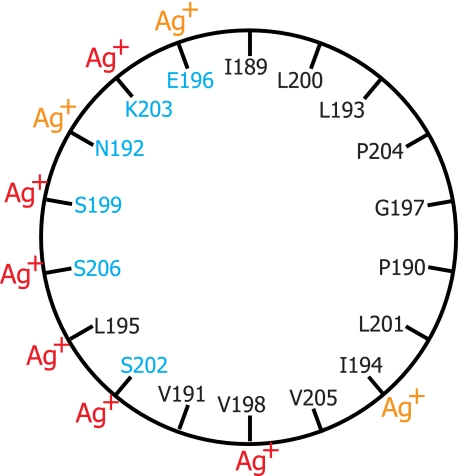
The Ag+-sensitive and cross-linkable region of loop 3–4 extending from Ile-189 to Lys-203 was suggested previously to pack near the cytoplasmic surface of TMH4 (38). In early models for subunit a, the region was thought to be transmembrane, and the semiconserved Glu-196 residue was once proposed to form a portion of the proton translocation pathway (39). The region also contains three sites leading to oligomycin resistance in mitochondria at positions equivalent to residues 195, 196, and 199 in E. coli (40–42). The other locus encoding oligomycin resistance in yeast mitochondrial subunit a maps to a region near His-245 in aTMH5 of E. coli (42). The other mutations giving rise to oligomycin or venturicidin resistance in yeast mitochondria encircle the equivalent of the Asp-61 residue in cTMH2 or lie in residues immediately opposite to Asp-61 in cTMH1 (43, 44). Venturicidin resistance mutations have also been mapped to residue 28 in cTMH1 of E. coli (45). Oligomycin is a hydrophobic and relatively planar, macrocyclic lactone with dimensions of ∼10–15 Å (46), and these observations collectively suggest that its binding site may extend from the surface of the membrane in the region of Glu-196 to the area around Asp-61 in the midst of the bilayer. If the region surrounding Glu-196 in loop 3–4 is helical, it would also be amphipathic (Fig. 8), and conceivably the Ag+-sensitive surface shown in Fig. 8 could interact directly with the Ag+-sensitive pocket at the cytoplasmic end of TMH5. A possible electrostatic protein interaction at the membrane surface is supported by the unique chemical reactivity of the aE196C mutant (12). In the mapping of exposed aqueous accessible loop residues (12), E196C was the only cytoplasmic or periplasmic Cys whose reactivity with membrane-impermeant maleimides was dependent upon treatment with high salt. The region is also resistant to labeling with propionylbiocytin-maleimide (37).
FIGURE 8.
Amphipathic characteristics and Ag+ sensitivity of residues in an α-helical wheel model for the C-terminal region of the 3–4 loop. Polar or charged wild type residues are shown in blue, and hydrophobic residues are shown in black. Ag+-sensitive Cys substitutions are highlighted with red Ag+ indicating >85% inhibition and orange Ag+ indicating 66–85% inhibition.
The observation that loop 1–2 residues V86C and M93C can be cross-linked with L195C provides insight into two heretofore unexplained suppressor mutations. Fraga et al. (47) isolated a collection of second site suppressor mutations that promoted function of the cA24D/D61G aspartate interchange mutant. Of the 23 isolates, 10 mapped to residues in cTMH2, and 10 mapped to residues in aTMH4. The remaining three isolates mapped to either residue 92 in loop 1–2 or residue 198 in loop 3–4 and, based upon their assumed position in topological models of the protein at that time, provided no insight into how the function of essential Asp in the middle of the membrane was enhanced. The cross-linking results reported here suggest that the Ag+-sensitive pockets on the cytoplasmic sides of TMHs 4 and 5 and in the 1–2 and 3–4 loops may pack in a single three-dimensional domain, which based upon the second site suppressor studies, could have important interactions with the essential Asp during ATP synthesis.
What possible role could the Ag+-sensitive regions of loops 1–2 and 3–4 play in F0 function? We suggest that the Ag+-sensitive regions of the loops may be gating entrance or exit to the cytoplasmic H+ half-channel leading to Asp-61. Specifically small movements in the loop regions may be coupled to a repositioning of TMHs 4 and 5 to alternatively open and close half-channels to the two sides of the membrane. Movement of the loop positions could be mechanically forced by contacts with the polar loop region of subunit c as it moves past subunit a during the rotation of the c ring. The polar loop could nudge the 3–4 loop to reposition itself such that the connecting TMH4 reoriented itself to allow H+ in the periplasmic half-channel to access Asp-61. Recall that the periplasmic half-channel is predicted to be at the center of a four-helix bundle, and access from there to the c ring surface would require a swiveling of helices at the TMH4/TMH5 interface (27, 28). Once subunit c has moved completely past the 3–4 loop region, the loop could relax, and the transmembrane domains could return to their alternate positions. In this orientation Arg-210 on TMH4 would move close to Asp-61 and drive its deprotonation by salt bridge formation. The proton released would exit via the cytoplasmic half-channel extending from Asn-214 and Arg-210 at the center of the membrane to the pocket of Ag+-sensitive residues at the cytoplasmic surface. Cys residues introduced into this region of TMH4 are both NEM- and Ag+-sensitive, and this aqueous pocket may extend to the NEM-sensitive V262C substitution at the cytoplasmic face of TMH5 (28).
In suggesting a gating role for the cytoplasmic loop regions of subunit a in regulating alternate access of H+ to Asp-61 from the two sides of the membrane, one is immediately reminded of several elegant, structurally defined examples in other membrane transport proteins. In considering the possible interaction of the 3–4 loop with deeply membrane-embedded regions of TMHs 4 and 5, one remembers the pore helix and selectivity filter in the subunits of the KscA potassium channel that dip into the transmembrane core and then reemerge at the extracellular surface of the membrane (48). The pore helix/selectivity filter region is strongly predicted to be an extracellular loop by hydropathy plot analysis. The sarco/endoplasmic reticulum P-type Ca2+-ATPase provides a perhaps classic example of a moving cytoplasmic domain driving TMH rearrangements to gate ion release (49). A rotational movement of the A (actuator) domain toward the P (phosphorylation) domain is required for dephosphorylation of the E2P intermediate, and the movement is concluded to force the Ca2+-occluding TMHs apart to release Ca2+ to the sarcoplasmic reticulum lumen. The cytoplasmic domains of a bacterial Mg2+ transporter are proposed to move together and dimerize on Mg2+ binding and in so doing change the angle of packing of transmembrane pore helices from the open to closed state to regulate/gate Mg2+ entry into the cytoplasm (50). Based upon crystal structures of the GltPh Na+/Asp co-transporter and related co-transporters, a swinging extracellular helical hairpin is proposed to occlude bound Na+ and substrate and serve as the closed gate prior to TMH movement and gating to open the binding site to the cytoplasm (51). Finally rotation of the four-helix coiled coil, extracellular linker domain in a wide variety of prokaryotic sensory receptors is proposed to be linked to rotation of a membrane-spanning helix to promote transmembrane signaling (52). We thus think that there are ample precedents to speculate that movements of the Ag+-sensitive cytoplasmic domains of subunit a may also provoke changes in the alignment of TMHs 4 and 5 to gate H+ access and exit from the Asp-61 H+ binding site in subunit c.
Supplementary Material
Acknowledgments
We thank Hun Sun Chung, Kelly A. G. Herold, and Nick van Gompel for assistance in some of the experiments.
This work was supported, in whole or in part, by National Institutes of Health Grant GM23105 from the United States Public Health Service. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains a supplemental table summarizing the properties of all Cys substitutions made in subunit a from residues 39–270, including both Ag+ and NEM sensitivity.
Footnotes
The abbreviations used are: TMH, transmembrane helix; ACMA, 9-amino-6-chloro-2-methoxyacridine; DABMI, 4-dimethylaminophenylazophenyl-4′-maleimide; MTS, methanethiosulfonate; M2M, 1,2-ethanediyl bis-MTS; M4M, 1,4-butanediyl bis-MTS; NEM, N-ethylmaleimide; PVDF, polyvinylidene difluoride.
References
- 1.Yoshida, M., Muneyuki, E., and Hisabori, T. (2001) Nat. Rev. Mol. Biol. 2 669–677 [DOI] [PubMed] [Google Scholar]
- 2.Capaldi, R. A., and Aggeler, R. (2002) Trends Biochem. Sci. 27 154–160 [DOI] [PubMed] [Google Scholar]
- 3.Dimroth, P., von Ballmoos, C., and Meier, T. (2006) EMBO Rep. 7 276–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Senior, A. E. (1988) Physiol. Rev. 68 177–231 [DOI] [PubMed] [Google Scholar]
- 5.Jiang, W., Hermolin, J., and Fillingame, R. H. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 4966–4971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitome, N., Suzuki, T., Hayashi, S., and Yoshida, M. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 12159–12164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meier, T., Polzer, P., Diederichs, K., Welte, W., and Dimroth, P. (2005) Science 308 659–662 [DOI] [PubMed] [Google Scholar]
- 8.Pogoryelov, D., Yu, J., Meier, T., Vonck, J., Dimroth, P., and Muller, D. J. (2005) EMBO Rep. 6 1040–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones, P. C., Jiang, W., and Fillingame, R. H. (1998) J. Biol. Chem. 273 17178–17185 [DOI] [PubMed] [Google Scholar]
- 10.Dmitriev, O. Y., Jones, P. C., and Fillingame, R. H. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 7785–7790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fillingame, R., Angevine, C., and Dmitriev, O. (2003) FEBS Lett. 555 29–34 [DOI] [PubMed] [Google Scholar]
- 12.Valiyaveetil, F. I., and Fillingame, R. H. (1998) J. Biol. Chem. 273 16241–16247 [DOI] [PubMed] [Google Scholar]
- 13.Long, J. C., Wang, S., and Vik, S. B. (1998) J. Biol. Chem. 273 16235–16240 [DOI] [PubMed] [Google Scholar]
- 14.Wada, T., Long, J. C., Zhang, D., and Vik, S. B. (1999) J. Biol. Chem. 274 17353–17357 [DOI] [PubMed] [Google Scholar]
- 15.Cain, B. D. (2000) J. Bioenerg. Biomembr. 32 365–371 [DOI] [PubMed] [Google Scholar]
- 16.Hatch, L. P., Cox, G. B., and Howitt, S. M. (1995) J. Biol. Chem. 270 29407–29412 [DOI] [PubMed] [Google Scholar]
- 17.Valiyaveetil, F. I., and Fillingame, R. H. (1997) J. Biol. Chem. 272 32635–32641 [DOI] [PubMed] [Google Scholar]
- 18.Jiang, W., and Fillingame, R. H. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 6607–6612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwem, B. E., and Fillingame, R. H. (2006) J. Biol. Chem. 281 37861–378670 [DOI] [PubMed] [Google Scholar]
- 20.Roberts, D. D., Lewis, S. D., Ballou, D. P., Olson, S. T., and Shafer, J. A. (1986) Biochemistry 25 5595–5601 [DOI] [PubMed] [Google Scholar]
- 21.Loo, T. W., and Clarke, D. M. (2001) J. Biol. Chem. 276 36877–36880 [DOI] [PubMed] [Google Scholar]
- 22.Mordoch, S. S., Granot, D., Lebendiker, M., and Schuldiner, S. (1999) J. Biol. Chem. 274 19480–19486 [DOI] [PubMed] [Google Scholar]
- 23.Tamura, N., Konishi, S., Iwaki, S., Kimura-Someya, T., Nada, S., and Yamaguchi, A. (2001) J. Biol. Chem. 276 20330–20339 [DOI] [PubMed] [Google Scholar]
- 24.Lu, Q., and Miller, C. (1995) Science 268 304–307 [DOI] [PubMed] [Google Scholar]
- 25.Li, J., Xu, Q., Cortes, D. M., Perozo, E., Laskey, A., and Karlin, A. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 11605–11610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Angevine, C. M., and Fillingame, R. H. (2003) J. Biol. Chem. 278 6066–6074 [DOI] [PubMed] [Google Scholar]
- 27.Angevine, C. M., Herold, K. A., and Fillingame, R. H. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 13179–13183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Angevine, C. M., Herold, K. A., Vincent, O. D., and Fillingame, R. H. (2007) J. Biol. Chem. 282 9001–9007 [DOI] [PubMed] [Google Scholar]
- 29.Kuo, P. H., Ketchum, C. J., and Nakamoto, R. K. (1998) FEBS Lett. 426 217–220 [DOI] [PubMed] [Google Scholar]
- 30.Barik, S. (1996) Methods Mol. Biol. 57 203–215 [DOI] [PubMed] [Google Scholar]
- 31.Walker, J. E., Saraste, M., and Gay, N. J. (1984) Biochim. Biophys. Acta 768 164–200 [DOI] [PubMed] [Google Scholar]
- 32.Mosher, M. E., White, L. K., Hermolin, J., and Fillingame, R. H. (1985) J. Biol. Chem. 260 4807–4814 [PubMed] [Google Scholar]
- 33.Fillingame, R. H. (1975) J. Bacteriol. 124 870–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laemmli, U. K. (1970) Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- 35.Towbin, H., Staehelin, T., and Gordon, J. (1979) Proc. Natl. Acad. Sci. U. S. A. 76 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hermolin, J., and Fillingame, R. H. (1995) J. Biol. Chem. 270 2815–2817 [DOI] [PubMed] [Google Scholar]
- 37.Zhang, D., and Vik, S. B. (2003) J. Biol. Chem. 278 12319–12324 [DOI] [PubMed] [Google Scholar]
- 38.Fillingame, R. H., Jiang, W., Dmitriev, P. C., and Jones, P. C. (2000) Biochim. Biophys. Acta 1458 387–403 [DOI] [PubMed] [Google Scholar]
- 39.Cox, G. B., Fimmel, A. L., Gibson, F., and Hatch, L. (1986) Biochim. Biophys. Acta 849 62–69 [DOI] [PubMed] [Google Scholar]
- 40.John, U. P., and Nagley, P. (1986) FEBS Lett. 207 79–83 [DOI] [PubMed] [Google Scholar]
- 41.Breen, G. A. M., Miller, D. L., Holmans, P. L., and Welch, G. (1986) J. Biol. Chem. 261 11680–11685 [PubMed] [Google Scholar]
- 42.Ray, M. K., Connerton, I. F., and Griffiths, D. E. (1988) Biochim. Biophys. Acta 951 213–219 [DOI] [PubMed] [Google Scholar]
- 43.Nagley, P., Hall, R. M., and Ooi, B. G. (1986) FEBS Lett. 195 159–163 [DOI] [PubMed] [Google Scholar]
- 44.Galinis, M., Mattoon, J. R., and Nagley, P. (1989) FEBS Lett. 249 333–336 [DOI] [PubMed] [Google Scholar]
- 45.Fillingame, R. H., Oldenburg, M., and Fraga, D. (1991) J. Biol. Chem. 266 20934–20939 [PubMed] [Google Scholar]
- 46.von Glehn, M., Norrestam, R., Kierkegaard, P., Maron, L., and Ernster, L. (1972) FEBS Lett. 20 267–269 [DOI] [PubMed] [Google Scholar]
- 47.Fraga, D., Hermolin, J., and Fillingame, R. H. (1994) J. Biol. Chem. 269 2562–2567 [PubMed] [Google Scholar]
- 48.Doyle, D. A., Cabral, J. M., Pfuetzner, R. A., Kuo, A., Gulbis, J. M., Cohen, S. L., Chair, B. T., and MacKinnon, R. (1998) Science 280 69–77 [DOI] [PubMed] [Google Scholar]
- 49.Olesen, C., Picard, M., Winther, A.-M. L., Gyrup, C., Morth, J. P., Oxvig, C., Moeller, J. V., and Nissen, P. (2007) Nature 450 1036–1042 [DOI] [PubMed] [Google Scholar]
- 50.Hattori, M., Tanaka, Y., Fukai, S., Ishitani, R., and Nureki, O. (2007) Nature 448 1072. [DOI] [PubMed] [Google Scholar]
- 51.Boudker, O., Ryan, R. M., Yernool, D., Shimamoto, K., and Gouaus, E. (2007) Nature 445 387–393 [DOI] [PubMed] [Google Scholar]
- 52.Hulko, M., Berndt, F., Gruber, M., Linder, J. U., Truffault, V., Schultz, A., Martin, J., Schultz, J. E., Lupas, A. N., and Coles, M. (2006) Cell 126 929–940 [DOI] [PubMed] [Google Scholar]
- 53.Dmitriev, O. Y., and Fillingame, R. H. (2004) J. Biomol. NMR 29 439–440 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.