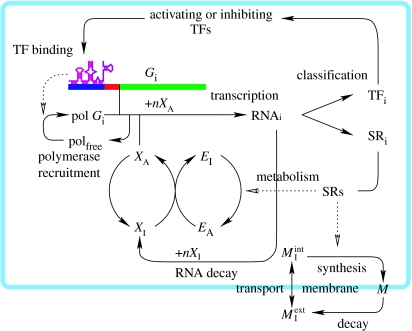
Figure 3.
A sketch of the chemical reaction network of the presented model. The RNA polymerase is assumed to be available in a fixed amount and recruited to the gene (green) promoter region (red) at a rate determined by the TFs bound to the URR (blue). The RNA transcription rate depends on the concentration of activated RNA building blocks (XA) and consumes nXA per RNA. The RNAs decay to inactivated components (XI), which are reactivated via consumption of activated, energy-rich metabolites (EA). The gene products are categorized into structural (SR) and gene regulatory (TF) RNAs. The SRs catalyse the activation of metabolites (EI) and the incorporation of membrane building blocks into the membrane (M). The internal pool of is coupled with the exterior pool via diffusion through the membrane. All the parameters for transcription factor binding to regulatory regions and the catalytic efficiencies of structural proteins are obtained by a mapping process (see text for details), and are therefore targets of evolution.