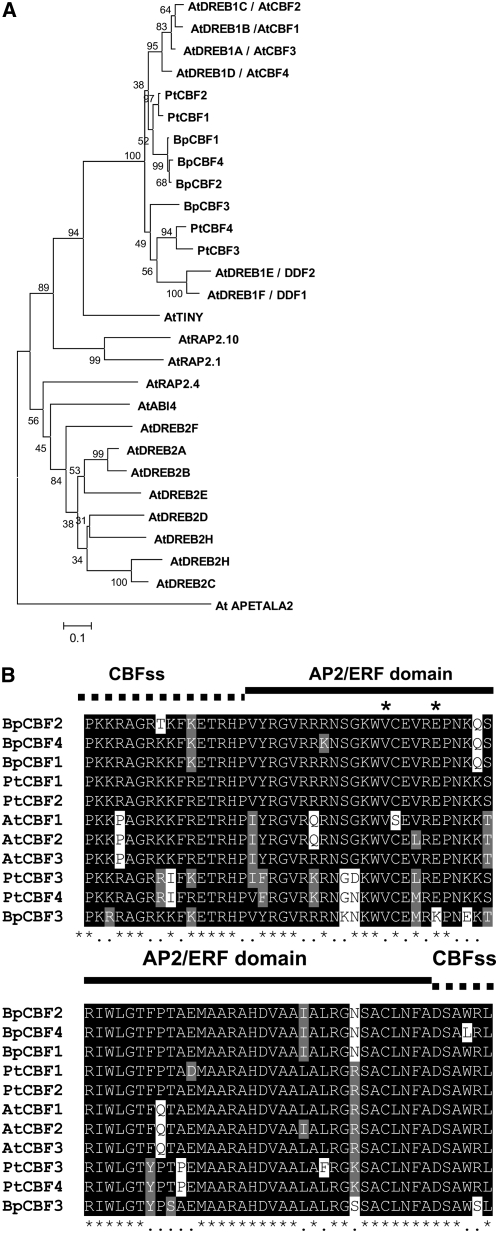
Figure 1.
Comparison of CBF/DREB1 protein family members of Arabidopsis, poplar, and birch. A, A phylogenetic analysis of birch and poplar CBF proteins and the Arabidopsis DREB family of proteins. Analysis, based on minimum evolution, was performed with the full-length protein sequences using the AP2 transcription factor as an out group. Alignment was made using four birch CBF proteins: BpCBF1 (ABP98987), BpCBF2 (ABP98988), BpCBF3 (ABP98989), and BpCBF4 (FG124897); four poplar CBF proteins: PtCBF1 (JGI666968), PtCBF2 (JGI346104), PtCBF3 (JGI548519), and PtCBF4 (JGI63608); and the Arabidopsis DREB family of proteins, which includes four CBF/DREB1 proteins: AtDREB1b/AtCBF1 (At4g25490), AtDREB1c/AtCBF2 (At4g25470), AtDREB1a/AtCBF3 (At4g25480), and AtDREB1d/AtCBF4 (At5g51990), as well as AtDREB1F/DDF1 (At1g12610), AtDREB1E/DDF2 (At1g63030), AtTINY (At5g25810), AtRAP2.10 (At4g36900), AtRAP2.1 (At1g46768), AtABI4 (At2g40220), AtRAP2.4 (At1g78080), AtDREB2D (At1g75490), AtDREB2G (At5g18450), AtDREB2F (At3g57600), AtDREB2E (At2g38340), AtDREB2A (At5g05410), AtDREB2B (At3g11020), AtDREB2H (At2g40350), AtDREB2C (At2g40340), and AtAP2 (At4g36920). CBF proteins were initially aligned using ClustalW and used to conduct phylogenetic analysis using MEGA version 4.1 software (http://www.megasoftware.net/). The phylogenetic tree was constructed using the neighbor-joining method on 1,000 bootstrap replications. Bootstrap percentages are shown on the dendrogram branch points. B, Alignment of AP2 and flanking CBF signature sequences of Arabidopsis, poplar, and birch CBF transcription factors. CBF signature sequences (CBFss) are marked with a dotted line, and the AP2/ERF domain is marked with a solid line. The 14th and 19th amino acids of AP2/ERF domain are marked with asterisks.