Figure 4.
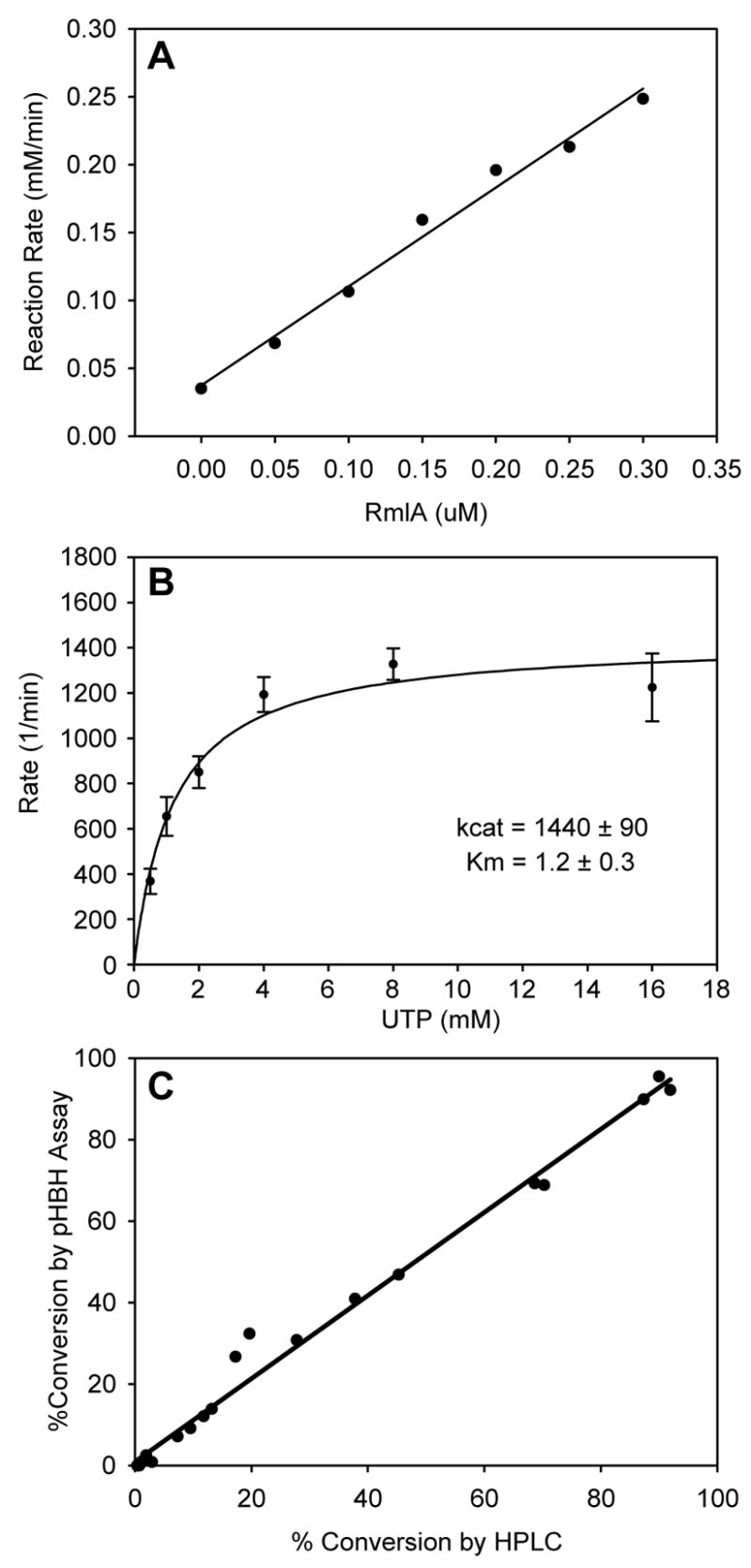
Validation of the phosphatase-assisted plate-based pHBH nucleotidyltransferase assay. (A) The observed rate of glucose-1-phosphate disappearance, under constant substrate concentrations (2 mM glucose and 2.5 mM UTP), plotted against enzyme concentration. (B) Determination of steady-state kinetics via the pHBH assay. Purified S. enterica RmlA (0.3 μM) was incubated with 2 mM glucose-1-phosphate and varying amounts of UTP. Aliquots were taken every 30s, and assayed by the plate-form pHBH assay. The rates of glucose-1-phosphate disappearance (corrected for the constant 0.3 μM enzyme concentration) versus UTP were fit to the Michaelis-Menten equation. (C) Comparison of activities determined by plate-form pHBH assay and HPLC. The activities of crude lysates from E. coli BL21(DE3) cells expressing RmlA variants representing a range of catalytic efficiencies (wild type, Q83A, Q83S, Q83D, Q83N and Q83E) were assessed by both by plate-form pHBH and standard HPLC methods at three different time points (2 mM glucose-1-phosphate, 2 mM UTP; reaction times - 10, 30 and 60 min). The results of the two sets of assays (pHBH versus HPLC) were plotted against one another (slope of line = 1.02, R2 = 0.9884).
