Abstract
Pathogens causing significant respiratory disease in growing pigs include Porcine reproductive and respiratory syndrome virus, Porcine circovirus 2, swine influenza virus, porcine respiratory coronavirus, Mycoplasma hyopneumoniae, and Bordetella bronchiseptica. The objective of this research was to characterize the respiratory excretion of these pathogens by acutely infected pigs. Pigs were inoculated under experimental conditions with 1 pathogen. Samples were collected from the upper respiratory tract and exhaled air. All pathogens were detected in swabs of the upper respiratory tract, but only M. hyopneumoniae and B. bronchiseptica were detected in expired air from individually sampled, acutely infected pigs. These findings suggest either that the acutely infected pigs did not aerosolize the viruses or that the quantity of virus excreted was below the detection threshold of current sampling or assay systems, or both, at the individual-pig level.
Résumé
Les agents pathogènes causant des maladies respiratoires chez les porcs en croissance incluent le virus du syndrome reproducteur et respiratoire porcin, le circovirus porcin de type 2, le virus de l’influenza porcin (SIV), le coronavirus respiratoire porcin, Mycoplasma hyopneumoniae et Bordetella bronchiseptica. La présente étude vise à caractériser l’excrétion de ces agents pathogènes dans les respirations de porcs souffrant d’infection aiguë. Des porcs ont été inoculés expérimentalement avec un de ces agents pathogènes. Des échantillons ont été prélevés au niveau du tractus respiratoire supérieur et des expirations respiratoires. Tous les agents pathogènes ont été détectés dans les voies respiratoires supérieures des porcs inoculés, mais seulement M. hyopneumoniae et B. bronchiseptica ont été détectés dans l’air expiré d’animaux individuels infectés de manière aiguë. Ces trouvailles suggèrent, soit que les porcs avec infection expérimentale aiguë n’aérosolisent pas PRRSV, PCV-2, SIV ou PRCV, ou que les quantités de virus excrétées sont inférieures à la sensibilité analytique (seuil de détection) des méthodes d’échantillonnage et/ou des méthodes d’analyse au niveau des individus.
(Traduit par Docteur Serge Messier)
Transmission from infected to susceptible animals via aerosols has been demonstrated under experimental conditions for Porcine reproductive and respiratory syndrome virus (PRRSV) (1,2), influenza virus (3), Porcine respiratory coronavirus (PRCoV) (4), Mycoplasma hyopneumoniae (5), and Bordetella bronchiseptica (1). In these studies, transmission between animals provided descriptive evidence that, under the specific conditions of the experiment, the pathogens a) were shed in the exhaled air of infected animals, b) remained airborne and infectious, and c) reached a susceptible animal in a dose sufficient to cause infection. Determining the quantity of pathogen aerosolized over time is a key step in the goal of understanding the parameters of aerosol transmission. Therefore, our objective was to quantify the excretion of pathogens [PRRSV, swine influenza virus (SIV), PRCoV, Porcine circovirus-2 (PCV-2), M. hyopneumoniae, and B. bronchiseptica] in air exhaled by acutely infected pigs over time after inoculation.
Two strains of PRRSV (ATCC VR-2332 and ATCC VR-2385; American Type Culture Collection, Manassas, Virginia, USA) were used. The viruses were propagated in MARC-145 monkey kidney cells (6). Pigs were inoculated intramuscularly with 1 mL of cell culture medium containing 1 × 102.0 50% tissue culture infectious dose (TCID50)/mL of VR-2332 PRRSV or intranasally with 2 mL of cell culture medium containing 1 × 105.2 TCID50/mL of VR-2385 PRRSV. A quantitative real-time reverse transcriptase polymerase chain reaction (RT-PCR) was used to detect PRRSV in impinger and swab samples (7).
Swine influenza virus [A/Swine/Minnesota/37866/1999 (H1N1) National Veterinary Service Laboratories, Ames, Iowa, USA] was propagated in Madin-Darby canine kidney cells (8). Pigs were inoculated intranasally with 3 mL of cell culture medium containing 1 × 106.3 TCID50/mL of SIV. A quantitative RT-PCR was used to detect SIV in impinger and swab samples (7).
A field strain of PRCoV was propagated in swine testicular cells (9). Pigs were inoculated intranasally with 4 mL of cell culture medium containing 1 × 106.0 TCID50/mL of PRCoV. A real-time multiplex RT-PCR designed to detect and differentiate Transmissible gastroenteritis virus (TGEV) and PRCoV was used to detect PRCoV in impinger samples. Primers and minor groove-binding (MGB) probes were based on nucleoprotein (N) and spike (S) protein gene sequences of TGEV or PRCoV available from the GenBank sequence database and manufactured by Integrated DNA Technologies (Coralville, Iowa, USA) and Applied Biosystems (Foster City, California, USA), respectively. Ribonucleic acid (RNA) was extracted from samples by means of the MagMAX Viral RNA Kit (Ambion, Austin, Texas, USA) according to the manufacturer’s protocol. The reverse-transcriptase polymerase chain reaction (RT-PCR) was carried out in an ABI7500 thermocycler (Applied Biosystems), 9600 emulation mode, with the QuantiTect Probe RT-PCR Kit (Qiagen, Valencia, California, USA) according to the manufacturer’s protocol. Running conditions were: 50°C for 30 min followed by 95°C for 30 min. Cycling was performed at 94°C for 15 s and then at 60°C for 60 s for a total of 40 cycles. Data acquisition was performed during the combined annealing/extension step at 60°C. The cutoff threshold cycle for positivity for each gene (S or N) was < 40. Samples positive only for the N gene were considered to be positive for PRCoV, wheras samples positive for both genes were considered to be positive for TGEV.
A field strain of PCV-2 (ISU-40895) was propagated on porcine kidney cells (10). Pigs were inoculated intranasally with 6 mL of cell culture medium containing 1 × 104.8 TCID50/mL of PCV-2. A PCR-based assay was used to detect PCV-2 in impinger and swab samples (11).
Mycoplasma hyopneumoniae strain 232, a derivative of M. hyopneumoniae strain 11, was propagated in Friis medium (12). Pigs were inoculated intratracheally with 10 mL of medium containing 1 × 104.0 color-changing units per milliliter. A PCR-based assay was used to detect M. hyopneumoniae in impinger and swab samples (13).
A field strain of B. bronchiseptica (strain KM22) was cultured on Bordet-Gengou agar and then suspended in phosphate-buffered saline (PBS) (8). Pigs were inoculated intranasally with 1 mL of PBS containing 1 × 106.0 colony-forming units of B. bronchiseptica. Bacterial isolation was used to detect B. bronchiseptica in impinger and swab samples (1).
For each pathogen, a group of pigs was inoculated under experimental conditions with 1 of the isolates described previously. Thereafter, oral swabs (for PRRSV detection), nasal swabs (for the detection of all the other pathogens), samples of air from the pigs, and samples of ambient room air were collected at regular intervals during the acute phase of the infection and assayed for the presence of the target pathogens (Table I).
Table I.
Schedule of aerosol and swab (nasal or oral) sampling to detect specific pathogens exhaled by inoculated pigs
| Sample type and number of days after inoculation
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pathogen targeted | Number of pigs | Agea (d) | 1 | 2 | 4 | 6 | 8 | 9 | 11 | 13 | 14 | 15 | 17 | 21 | 23 | 28 |
| PRRSVb | 20 | 21 | AE | ns | AE | AEO | AEO | ns | AEO | AEO | ns | AEO | ns | ns | ns | ns |
| PRRSVc | 6 | 56 | AEO | ns | AEO | AEO | AEO | ns | AEO | AEO | ns | AEO | ns | ns | ns | ns |
| PCV-2d | 4 | 63 | AEN | AEN | ns | AEN | AEN | ns | ns | ns | AEN | ns | ns | ns | ns | ns |
| PRCoVe | 4 | 24 | ns | AEN | AEN | AEN | AEN | AEN | ns | ns | ns | ns | ns | ns | ns | ns |
| SIVf | 4 | 24 | ns | AEN | AEN | ns | AEN | AEN | ns | ns | ns | ns | ns | ns | ns | ns |
| M. hyopneumoniaeg | 4 | 35 | AEN | ns | AEN | AEN | AEN | ns | AEN | AEN | ns | AEN | AEN | AEN | AEN | AEN |
| B. bronchisepticah | 4 | 24 | ns | AEN | AEN | AEN | AEN | AEN | ns | ns | ns | ns | ns | ns | ns | ns |
B. — Bordetella; M.— Mycoplasma; A — aerosol sample collected in glass impinger; E — environmental (room air) sample; ns — no sample; O — oral swab; N — nasal swab.
Age at time of inoculation.
PRRSV — Porcine reproductive and respiratory syndrome virus Isolate ATCC VR-2332.
Isolate, ATCC VR-2385.
PCV-2 — Porcine circovirus Isolate ISU-40895.
PRCoV — Porcine respiratory coronavirus field isolate.
SIV — Swine influenza virus A/Swine/Minnesota/37866/1999 (H1N1).
Mycoplasma hyopneumoniae Strain 232.
Bordetella bronchiseptica Strain KM22.
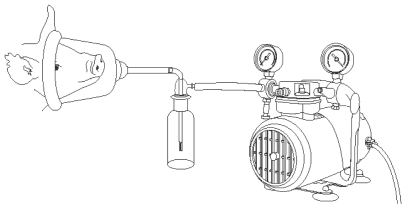
Samples of expired air were collected from unanesthetized pigs for 5 min. Each pig was held, and the snout placed into a large canine surgical mask (model SurgiVet, 32393B1; Waukesha, Wisconsin, USA) connected to an AGI-30 glass impinger (Ace Glass, Vineland, New Jersey, USA) with clear tubing (Fisher Scientific, Hampton, New Hampshire, USA) (Figure 1). Each impinger contained 20 mL of sterile PBS (pH 7.4) (1 X) (Invitrogen, Carlsbad, California, USA) collection fluid. The impingers were operated at a vacuum pressure of less than half an atmosphere with the use of oil-less pumps (model S413801; Fisher Scientific). This ensured a constant sampling flow rate of 12.5 L/min. The vacuum pressure was monitored constantly by means of a vacuum pressure gauge (model G-S4LM20-VAC-100; Cato Western, Tucson, Arizona, USA). To avoid negative pressure on the pig and facilitate the flow of air from the pig to the impinger, 4 holes (0.158 cm diameter) were made in the sides of the masks. After sampling, aliquots of the impinger collection fluid were placed in snap-cap tubes (model 14-956-1B; Fisher Scientific).
Figure 1.
Diagrammatic representation of system used to collect respiratory samples from pigs.
Nasal or oral samples were collected by means of sterile polyester swabs (Fisher Scientific). Nasal samples were collected by inserting and rotating a swab in the nares. Oral samples were collected by dragging and rotating the swab along the gingival crevicular surface of the oral cavity. All swabs were immediately placed in snap-cap tubes containing 2 mL of sterile PBS.
All samples were stored frozen at −80°C until tested. After completion of the study, the samples were completely randomized, relabeled, and then assayed as a block for the presence of the target pathogens. A sample was considered positive if the target pathogen was detected by the specified assay (Table II).
Table II.
Summary of methods of inoculation and detection of the targeted pathogens
| Pathogena | Strain | Route | Volume | Dose | Assay | Laboratory |
|---|---|---|---|---|---|---|
| PRRSV (7) | VR-2332 | IM | 1.0 mL | 102.0 TCID50/mL | RT-PCR | ISU |
| PRRSV (7) | VR-2385 | IN | 2.0 mL | 105.2 TCID50/mL | RT-PCR | ISU |
| SIV (7) | H1N1 | IN | 3.0 mL | 106.3 TCID50/mL | RT-PCR | ISU |
| PRCoV | Field | IN | 4.0 mL | 106.0 TCID50/mL | PCR | ISU |
| PCV-2 (11) | ISU-40895 | IN | 6.0 mL | 104.8 TCID50/mL | PCR | SDSU |
| M. hyopneumoniae (13) | 232 | IT | 10 mL | 104.0 CCU/mL | PCR | SDSU |
| B. bronchiseptica (1) | KM22 | IN | 1.0 mL | 106.0 CFU/mL | Isolation | NADC |
IM — intramuscular; IN — intranasal; IT — intratracheal; TCID50 — 50% tissue culture infectious dose; CCU — color changing units; CFU — colony-forming units; RT — reverse transcriptase; PCR — polymerase chain reaction; ISU — Iowa State University; SDSU — South Dakota State University; NADC — National Animal Disease Center.
Isolates and strains as in Table I. References ( ) are to previous methodologic descriptions.
Swab samples (oral or nasal) demonstrated that all the targeted pathogens were present at detectable levels in the upper respiratory tract (Table III). The day of initial detection, frequency, and proportion of positive samples among the swabs varied by pathogen. All pathogens except PCV-2 and M. hyopneumoniae were first detected in 1 or more pigs from 2 to 4 days after inoculation; in contrast, PCV-2 and M. hyopneumoniae were not detected until day 14. The highest rate of positivity among the nasal swabs (20/20) was for B. bronchiseptica, the lowest rate (3/20) for PCV-2 over the course of sampling.
Table III.
Assay results
| Number of positive samples; days after inoculation
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pathogen | Sample type | 1 | 2 | 4 | 6 | 8 | 9 | 11 | 13 | 14 | 15 | 17 | 21 | 23 | 28 | Totals |
| PRRSV VR-2332 | Aerosol | ns | ns | ns | 0 | 0 | ns | 0 | 0 | ns | 0 | ns | ns | ns | ns | 0/100 |
| PRRSV VR-2385 | Aerosol | 0 | ns | 0 | ns | 0 | ns | 0 | 0 | ns | 0 | ns | ns | ns | ns | 0/36 |
| PCV-2 | Aerosol | 0 | 0 | ns | 0 | 0 | ns | ns | ns | 0 | ns | ns | ns | ns | ns | 0/20 |
| PRCoV | Aerosol | ns | 0 | 0 | 0 | 0 | 0 | ns | ns | ns | ns | ns | ns | ns | ns | 0/20 |
| SIV | Aerosol | ns | 0 | 0 | ns | 0 | 0 | ns | ns | ns | ns | ns | ns | ns | ns | 0/16 |
| M. hyopneumoniae | Aerosol | 0 | ns | 0 | 0 | 1 | ns | 0 | ns | 0 | 0 | 1 | 1 | 0 | 1 | 4/44 |
| B. bronchiseptica | Aerosol | ns | 0 | 0 | 2 | 0 | 0 | ns | ns | ns | ns | ns | ns | ns | ns | 2/20 |
| PRRSV VR-2332 | Oral swab | ns | ns | ns | 10 | 5 | ns | 6 | 6 | ns | 2 | ns | ns | ns | ns | 29/100 |
| PRRSV VR-2385 | Oral swab | 0 | ns | 4 | ns | 1 | ns | 2 | 0 | ns | 0 | ns | ns | ns | ns | 7/36 |
| PCV-2 | Nasal swab | 0 | 0 | ns | 0 | 0 | ns | ns | ns | 3 | ns | ns | ns | ns | ns | 3/20 |
| PRCV | Nasal swab | ns | 3 | 2 | 2 | 0 | 0 | ns | ns | ns | ns | ns | ns | ns | ns | 7/20 |
| SIV | Nasal swab | ns | 4 | 4 | ns | 0 | 0 | ns | ns | ns | ns | ns | ns | ns | ns | 8/16 |
| M. hyopneumoniae | Nasal swab | 0 | ns | 0 | 0 | 0 | ns | 0 | ns | 2 | 2 | 4 | 4 | 3 | 4 | 19/44 |
| B. bronchiseptica | Nasal swab | ns | 4 | 4 | 4 | 4 | 4 | ns | ns | ns | ns | ns | ns | ns | ns | 20/20 |
The aerosol results did not generally reflect the swab results (Table III). None of the air samples were positive for the targeted viruses, whereas both bacteria were detected: M. hyopneumoniae was recovered from respiratory samples from 4 different pigs on days 8, 17, 21, and 28 after inoculation, for a positivity rate of 9% (4 of 44 samples); B. bronchiseptica was recovered from respiratory samples from 2 of 4 pigs on day 6 for a positivity rate of 10% (2 of 20 samples). All ambient room air samples were negative for the targeted pathogens on all sampling days.
The objective of this research was to quantify pathogens in air exhaled from acutely infected pigs. The detection and recovery of B. bronchiseptica and M. hyopneumoniae agree with earlier reports based on air samples collected from infected pigs (5,14,15). The negative results for viral pathogens conflict with those from descriptive studies reporting airborne transmission of PRRSV (1), PRCoV (4), and influenza virus (3) between animals and those from quantitative studies reporting on exhaled PRRSV (15). However, these results agree with negative results for the detection of airborne PRRSV from infected pigs with sampling systems similar to that described herein (1,5). The experimental design of the previous studies differed from our design in that samples were collected from the air space in which groups of pigs were housed rather than from individual pigs.
Overall, these data indicate that the viral pathogens were either not present in exhaled air or were present at levels below the analytical sensitivity of the sampling and detection procedures. Sample collection time, analytical sensitivity of the detection system, and isolate pathogenicity may have contributed to the negative results (2,15).
Sample collection time may contribute substantially to the discordance between the results herein and those of animal transmission studies. Essentially, susceptible animals function as continuous in-vivo monitors of the presence of airborne infectious agents, thus optimizing the likelihood of “detection.” In contrast, in-vitro air samples are collected for a brief interval. Maximum sample collection time is mandated by the physical design of the sampler, but prolonged impingement is generally not practical because of desiccation of the sampling medium, physical destruction of targets, or re-entrainment (re-aerosolization) of pathogens.
Impinger performance is known to be affected by a number of other factors, including collection medium composition, sampler type, sampling time, sampler flow rate (7,16,17), collection efficiency, and particle size (18). The sampling and detection systems used in this study had previously been shown to be capable of detecting 1 × 101.1 TCID50 of PRRSV and 1 × 101.4 TCID50 of SIV excreted over a 5-min sampling period (19). Therefore, if PRRSV and SIV were aerosolized, the concentrations were below those of the impinger’s analytic sensitivity.
Overall, the results of this study confirm the findings of Cho et al (15) that airborne pathogens are generally aerosolized by individual pigs in minute quantities, which makes detection and quantification by current sampling systems difficult. Alternative approaches, such as novel collection methods, sampling of groups of pigs in defined air spaces, and technical improvements in air samplers, may be required to accurately estimate excretion rates at the individual-pig level. An important consideration with approaches that do not sample the individual pig is the underestimation of excretion rates due to pathogen dispersion and sedimentation. Regardless of the technical challenge, probabilistic models for within-site and between-site transmission of airborne pathogens await quantitative measures of the aerosol excretion of pathogens by pigs.
Acknowledgments
This project was funded by Check-Off Dollars through the National Pork Board. The authors thank Dr. Patrick Halbur, Iowa State University, for his valuable collaboration. The authors are grateful to the Virology Staff at the Veterinary Diagnostic Laboratory, College of Veterinary Medicine, Iowa State University, and the Animal Disease and Research Diagnostic Laboratory staff at South Dakota State University for excellent diagnostic testing.
References
- 1.Brockmeier SL, Lager KM. Experimental airborne transmission of porcine reproductive and respiratory syndrome virus and Bordetella bronchiseptica. Vet Microbiol. 2002;89:267–275. doi: 10.1016/s0378-1135(02)00204-3. [DOI] [PubMed] [Google Scholar]
- 2.Cho JG, Deen J, Dee SA. Influence of isolate pathogenicity on the aerosol transmission of Porcine reproductive and respiratory syndrome virus. Can J Vet Res. 2007;71:23–27. [PMC free article] [PubMed] [Google Scholar]
- 3.Schulman JL, Khakpour M, Kilbourne ED. Protective effects of specific immunity to viral neuraminidase on influenza virus infection of mice. J Virol. 1968;2:778–786. doi: 10.1128/jvi.2.8.778-786.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourgueil E, Hutet E, Cariolet R, Vannier P. Experimental infection of pigs with the porcine respiratory coronavirus (PRCV): measure of viral excretion. Vet Microbiol. 1992;31:11–18. doi: 10.1016/0378-1135(92)90136-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fano E, Pijoan C, Dee S. Evaluation of the aerosol transmission of a mixed infection of Mycoplasma hyopneumoniae and porcine reproductive and respiratory syndrome virus. Vet Rec. 2005;157:105–108. doi: 10.1136/vr.157.4.105. [DOI] [PubMed] [Google Scholar]
- 6.Kim HS, Kwang J, Yoon IJ, Joo HS, Frey ML. Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in a homogeneous subpopulation of MA-104 cells. Arch Virol. 1993;133:477–483. doi: 10.1007/BF01313785. [DOI] [PubMed] [Google Scholar]
- 7.Hermann JR, Hoff SJ, Yoon KJ, Burkhardt AC, Evans RB, Zimmerman JJ. Optimization of a sampling system for recovery and detection of airborne porcine reproductive and respiratory syndrome virus and swine influenza virus. Appl Environ Microbiol. 2006;72:4811–4818. doi: 10.1128/AEM.00472-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clavijo A, Tresnan DB, Jolie R, Zhou EM. Comparison of embryonated chicken eggs with MDCK cell culture for the isolation of swine influenza virus. Can J Vet Res. 2002;66:117–121. [PMC free article] [PubMed] [Google Scholar]
- 9.Brockmeier SL, Loving CL, Nicholson TL, Palmer MV. Coinfection of pigs with porcine respiratory coronavirus and Bordetella bronchiseptica. Vet Microbiol. 2008;128:36–47. doi: 10.1016/j.vetmic.2007.09.025. Epub 2007 Oct. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fenaux M, Halbur PG, Haqshenas G, et al. Cloned genomic DNA of type 2 porcine circovirus is infectious when injected directly into the liver and lymph nodes of pigs: Characterization of clinical disease, virus distribution, and pathologic lesions. J Virol. 2002;76:541–551. doi: 10.1128/JVI.76.2.541-551.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larochelle R, Antaya M, Morin M, Magar R. Typing of porcine circovirus in clinical specimens by multiplex PCR. J Virol Methods. 1999;80:69–75. doi: 10.1016/s0166-0934(99)00032-4. [DOI] [PubMed] [Google Scholar]
- 12.Ross RF, Zimmermann-Erickson BJ, Young TF. Characteristics of protective activity of Mycoplasma hyopneumoniae vaccine. Am J Vet Res. 1984;45:1899–1905. [PubMed] [Google Scholar]
- 13.Ahrens P, Friis NF. Identification of Mycoplasma hyopneumoniae with a DNA probe. Lett Appl Microbiol. 1991;12:249–253. doi: 10.1111/j.1472-765x.1991.tb00551.x. [DOI] [PubMed] [Google Scholar]
- 14.Stehmann R, Mehlhorn G, Neuparth V. [Characterization of strains of Bordetella bronchiseptica isolated from animal housing air and of the airborne infection pressure proceeding from them] Dtsch Tierarztl Wochenschr. 1991;98:448–450. [PubMed] [Google Scholar]
- 15.Cho JG, Dee SA, Deen J, et al. The impact of animal age, bacterial coinfection, and isolate pathogenicity on the shedding of Porcine reproductive and respiratory syndrome virus in aerosols from experimentally infected pigs. Can J Vet Res. 2006;70:297–301. [PMC free article] [PubMed] [Google Scholar]
- 16.Trincado C, Dee S, Jacobson L, Otake S, Rossow K, Pijoan C. Attempts to transmit porcine reproductive and respiratory syndrome virus by aerosols under controlled field conditions. Vet Rec. 2004;154:294–297. doi: 10.1136/vr.154.10.294. [DOI] [PubMed] [Google Scholar]
- 17.Juozaitis A, Willeke K, Grinshpun SA, Donnelly J. Impaction onto a glass slide or agar versus impingement into a liquid for the collection and recovery of airborne microorganisms. Appl Environ Microbiol. 1994;60:861–870. doi: 10.1128/aem.60.3.861-870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin X, Willeke K, Ulevicius V, Grinshpun SA. Effect of sampling time on the collection efficiency of all-glass impingers. Am Ind Hyg Assoc J. 1997;58:480–488. [Google Scholar]
- 19.Hermann JR, Yoon K-J, Hoff SJ, Zimmerman JJ. Analytical sensitivity of air samplers detecting airborne Porcine reproductive and respiratory syndrome virus and swine influenza virus from a uniform point source exposure. Can J Vet Res. 2008 accepted for publication. [PMC free article] [PubMed] [Google Scholar]