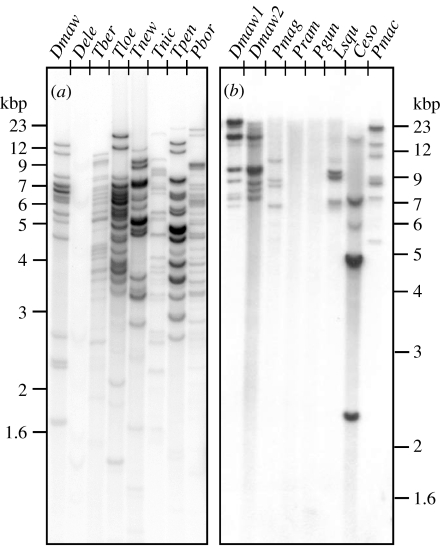
Figure 3.
Genomic DNA blots of notothenioid taxa hybridized with radio-labelled probe derived from a Notothenia coriiceps AFGP polyprotein gene. Genomic DNA was digested with (a) TaqI or (b) EcoRI, both known not to cleave within AFGP polyprotein coding sequences, thus each hybridized band represents one or more AFGP genes. (a) D. eleginoides (Dele) shows nearly undetectable hybridization signal indicating highly mutated or absence of AFGP sequences in its genome, in stark contrast to its fully AFGP-fortified sister species D. mawsoni (Dmaw), and related nototheniid relatives Trematomus bernacchii (Tber), T. loennbergii (Tloe), T. newnesi (Tnew), T. nicolai (Tnic), T. pennelli (Tpen) and Pagothenia borchgrevinki (Pbor). (b) The DNA of the two Patagonotothen species, P. ramsayi (Pram) from South America and P. guntheri (Pgun) from Shag Rocks, Antarctic Peninsula showed no hybridization, indicative of absence of functional AFGP sequences, in contrast to other nototheniids D. mawsoni (Dmaw1, Dmaw2), Paranotothenia magellanica (Pmag) and Lepidonotothen squamifrons (Lsqu). Like most of the Patagonotothen species, P. magellanica and the Channichthyid icefish Champsocephalus esox occur in South American waters, but show presence of AFGP sequences in their DNA, consistent with a species evolutionary origin in the Antarctic. Pmac is the icefish Pagetopsis macropterus.