Figure 4. ChIP Assay for Caveolin-1 Promoter.
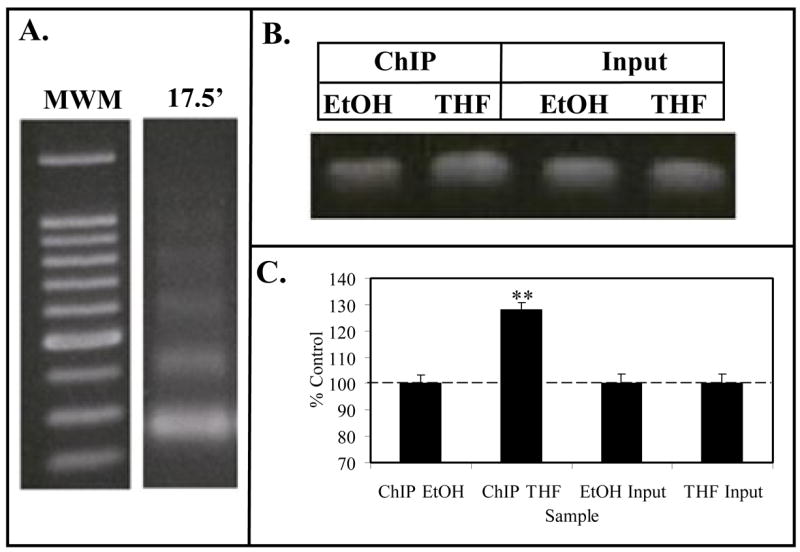
Panel A. Formaldehyde fixed MCF-7 cell chromatin was digested (17.5′) with Enzymatic Shearing Cocktail (Active Motif). Following cross-link reversal and proteinase K treatment, sheared DNA was electrophoresed on 1% agarose and stained with ethidium bromide. The gel contains a 100 bp molecular weight ladder (MWM), and DNA that yielded 100–200 bp fragments suitable for ChIP assays. Panel B (ChIP Assay). Following the immunoprecipitation chromatin from EtOH (controls) or THF-diol treated MCF-7 cells, the DNA was subjected to PCR analysis (36 cycles) with primers for the CAV1 promoter. PCR products from the Input DNA (1:10 dilution) are shown as additional controls. Results from 3 PCR reactions (as in panel B) for EtOH controls or THF-diol treated cells were quantified with UN-SCAN-IT (Silk Scientific Software), normalized to Input DNA and analyzed with Instat (panel C).
