Abstract
Conversion of the cellular prion protein (PrPC) into its altered conformation, PrPSc, is believed to be the major cause of prion diseases. Although PrP is the only identified agent for these diseases, there is increasing evidence that other molecules can modulate the conversion. We have found that interaction of PrP with double-stranded DNA leads to a protein with higher β-sheet content and characteristics similar to those of PrPSc. RNA molecules can also interact with PrP and potentially modulate PrPC to PrPSc conversion or even bind differentially to both PrP isoforms. Here, we investigated the interaction of recombinant murine PrP with synthetic RNA sequences and with total RNA extracted from cultured neuroblastoma cells (N2aRNA). We found that PrP interacts with N2aRNA with nanomolar affinity, aggregates upon this interaction, and forms species partially resistant to proteolysis. RNA does not bind to N-terminal deletion mutants of PrP, indicating that the N-terminal region is important for this process. Cell viability assays showed that only the N2aRNA extract induces PrP-RNA aggregates that can alter the homeostasis of cultured cells. Small RNAs bound to PrP give rise to nontoxic small oligomers. Nuclear magnetic resonance measurements of the PrP-RNA complex revealed structural changes in PrP, but most of its native fold is maintained. These results indicate that there is selectivity in the species generated by interaction with different molecules of RNA. The catalytic effect of RNA on the PrPC→PrPSc conversion depends on the RNA sequence, and small RNA molecules may exert a protective effect.
Prion diseases can be infectious, sporadic, or inherited (1). Regardless of their origin, they are related to modifications of a ubiquitous membrane-anchored protein, the prion protein (PrP).3 Through a poorly understood process, the cellular PrP isoform (PrPC), an α-helix-rich protein, undergoes a profound conformational change, acquiring higher β-sheet content; the latter isoform is known as PrPSc (Sc from scrapie) and is the only known component of the infectious prion particle (1–4).
The protein-only hypothesis postulates that PrPSc “multiplies” by catalyzing the conversion of PrPC into a likeness of itself, thus becoming responsible for its own propagation (5). This hypothesis is based strongly on the fact that PrP knock-out mice are resistant to prion infection, suggesting that endogenous PrP is necessary for prion propagation and infection (6). It was also suggested, however, that an additional unknown factor could influence the PrPC to PrPSc conversion (7–10). This molecule would act by lowering the free energy barrier between PrPC and PrPSc and triggering conversion (11, 12). In this field, a great number of biological macromolecules have emerged as candidates for conversion catalysts. Cellular adhesion molecules, nucleic acids (NAs), basal membrane molecules, and sulfated glycans, among other biological macromolecules, have been reported to interact with PrPC and to induce its conversion into a β-sheet-rich structure similar to the infectious prion protein (8, 10, 13, 14).
Our previous findings have demonstrated that PrP interacts with nucleic acids in vitro, binding small sequences of double-stranded DNA, acquiring β-sheet secondary structure as revealed by spectroscopic measurements and presenting some PrPSc-like characteristics (8, 15). It has also been shown that PrP interaction with nucleic acids can lead to partial unfolding of prion protein, triggering formation of an amyloid-like structure resistant to proteinase K digestion (8, 16–18), depending on the PrP-NA molar ratio.
The interaction of prion protein with ribonucleic acid has also been reported. PrPC interacts with mammalian RNA preparations, acquiring resistance to protease digestion in vitro (17, 19, 20). Highly structured RNAs can also convert human PrPC into PrPSc-like forms (21), and some RNA aptamers bind with high specificity to the disease-associated PrP conformation (22, 23). It has also been reported that the N terminus of the protein is important for the interaction with nucleic acids, because mutants lacking different portions of this region presented lower or no affinity for some RNA aptamers (24, 25), and DNA binding was obtained for a rPrP(23–144) construction (26).
All of these findings suggest that nucleic acids play a role in prion diseases. Based on these results, we have proposed a complement for the “protein-only” hypothesis in which nucleic acids can act by serving as catalysts and/or molecular chaperones in the conversion, not by encoding genetic information (8, 10, 13). More recently, Prusiner and co-workers (27) found that the binding of single-stranded DNA thioaptamers to PrP occurs on at least two different sites on the protein. Selection against recombinant Syrian hamster PrP 90–231 (recSHaPrP) identified a 12-base consensus sequence within a series of 20 thioaptamers, all of which consist of 40 bases, and one thioaptamer bound to recSHaPrP with extremely high affinity (0.58 nm) (27). The potential therapeutic use of these molecules against prion diseases reinforces the importance of understanding the interaction of PrP protein with different nucleic acids at the molecular level.
Besides the prion structural puzzle and the mystery involving the mechanisms of conversion from PrPC into PrPSc, many questions remain unanswered regarding the toxicity of prion aggregates. In addition to amyloid fibers and unstructured deposits of misfolded protein found in infected brains, smaller oligomers have been reported to be the toxic species for several amyloidogenic diseases, including transthyretin amyloidoses, Alzheimer, and Parkinson diseases (28–30). A series of PrP aggregates, oligomers, and fibers generated in vitro have been tested for toxicity in cultured cells and hamsters. In both cases, the aggregates were toxic, leading to cell death in culture and to neurodegeneration and spongiosis in hamster brains (31, 32). Moreover, it cannot be completely excluded that a small oligonucleotide may be present in PrP aggregates and account for prion cytotoxicity, as previously proposed (33).
Here we report experimental data on the interaction between murine recombinant prion protein (rPrP23–231) and RNA. We used total RNA extracted from cultured cells and small synthetic oligonucleotides, focusing on PrP secondary and tertiary structure changes. We have also investigated the role of the N-terminal PrP region in RNA binding using two mutants lacking N-terminal domains and evaluating the toxicity of the PrP-RNA complex in cultured mouse neuroblastoma cells (N2a). The recombinant prion protein (rPrP) deletion mutants investigated were rPrPΔ51–90 and rPrPΔ32–121, the former lacking residues 51–90, which is the copper-binding region, and the latter lacking residues 32–121, the majority of the PrP N terminus (34). We show that RNA extracted from N2a cells (N2aRNA) induces a loss of α-helical secondary structure and triggers aggregation of rPrP23–231 but has no effect on the rPrP-lacking portions of the N-terminal region. Nuclear magnetic resonance experiments reveal, however, that full-length rPrP partially recovers its native fold 3 days after RNA addition, but the changes observed in heteronuclear single quantum coherence (HSQC) spectrum suggest that RNA binding induces changes in PrP structure. We also find that aggregates generated from PrP-N2aRNA interaction are toxic to cultured N2a cells, which suggests that RNA molecules are potential candidates for catalyzing the PrPC to PrPSc conversion in vivo.
EXPERIMENTAL PROCEDURES
Construction, Expression, and Purification of Recombinant Prion Proteins—Construction of rPrP N-terminal deletion mutants rPrPΔ51–90 and rPrPΔ32–121, heterologous expression in Escherichia coli, and further purification of the constructs and full-length rPrP by high affinity column refolding followed previously described protocols (8, 34, 35).
rPrP23–231, rPrPΔ31–121, and rPrPΔ51–90 Labeling with Fluorescein Isothiocyanate—All of the rPrPs were labeled with amino-reactive fluorescein isothiocyanate (FITC) for fluorescence anisotropy assays. For the labeling reaction, rPrPs were dissolved in 0.1 m sodium bicarbonate buffer at pH 8.0 to a final concentration of 5–10 mg/ml and incubated with FITC (final concentration, 0.5–1.0 mg/ml) for 1 h at room temperature. After incubation, the FITC-rPrP23–231 conjugate (FITC-rPrP) was separated from the free dye by gel filtration using a PD-10 desalting column (Amersham Biosciences) equilibrated with Tris buffer (50 mm, pH 7.4) containing 100 mm NaCl. After elution, concentration of the labeled protein and the efficiency of labeling were determined based on molar extinction coefficients of rPrP23–231 (63,495 cm-1 m-1 at 280 nm), rPrPΔ32–121 (33,015 cm-1 m-1 at 280 nm), rPrPΔ51–90 (41,495 cm-1 m-1 at 280 nm), and FITC (68,000 cm-1 m-1 at 494 nm). FITC-rPrPs were stored at -20 °C and protected from light.
Ribonucleic Acid Samples—Total RNA from cultured neuroblastoma (N2a) cells (N2aRNA) (36) was extracted using the RNeasy Midikit (Qiagen) and the RNAspin Mini isolation kit (GE Healthcare, Milwaukee, WI). Other cell lineages used in this work are listed in the supplemental data. High pressure liquid chromatography-purified synthetic single-stranded RNA fragment 43–59 of SAF93 aptamer (SAF9343–59) (22) and opRNA, a random 18-mer sequence, derived from a translational operator of MS2 bacteriophage (37) were obtained from Integrated DNA Technologies, Inc. (Coralville, IA). Nucleotide sequences were: SAF9343–59, 5′-GGA UGC AAU CUC CAU CCC-3′; and opRNA, 5′-AAA CAU GGG UUC CCA UGU-3′. Synthetic RNA samples were maintained lyophilized at -20 °C and used in RNase-free water.
Reagents and Protein Samples—All of the reagents used were of analytical grade. Protein concentration was 1.0 μm (0.023 mg/ml for rPrP23–231, 0.018 mg/ml for rPrPΔ51–90, and 0.014 mg/ml for rPrPΔ32–121) for light scattering (LS) and fluorescence measurements, 30 μm (0.69 mg/ml for rPrP23–231, 0.57 mg/ml for rPrPΔ51–90, and 0.42 mg/ml for rPrPΔ32–121) for CD assays, and 50 nm (1.15 μg/ml for rPrP23–231, 0.9 μg/ml for rPrPΔ51–90, and 0.7 μg/ml for rPrPΔ32–121) for fluorescence anisotropy assays. All of the experiments were performed in 50 mm Tris buffer containing 100 mm NaCl, pH 7.4. All of the figures presented in this work are representative of at least three experiments.
Spectroscopic Measurements—LS, fluorescence anisotropy, and fluorescence emission were recorded on an ISSPC1 fluorometer (ISS, Champaign, IL). LS at 90 ° was measured illuminating the samples at 320 nm and collecting LS from 300 to 340 nm. Tryptophan fluorescence of rPrP23–231, rPrPΔ51–90, and rPrPΔ32–121 was measured by exciting the samples at 280 nm and collecting the emission from 300 to 420 nm. For anisotropy measurements, the samples were excited at 490 nm, and the emission was observed through a 3–69 filter.
Far-UV Circular Dichroism—CD spectra were recorded in a Jasco J-715 spectropolarimeter (Jasco Corporation, Tokyo, Japan) at 25 °C with circular 0.10-mm-pathlength cells. Buffer spectra were subtracted from each sample spectrum, and traces were collected with four accumulations each.
NMR Spectroscopy—NMR spectra were collected at 298 K with a Bruker Avance 800-MHz spectrometer equipped with gradient triple resonance probes. The spectra were processed using TOPSPIN 2.1 (Bruker) and analyzed with CARA 1.8 (38). Two-dimensional [15N,1H]HSQCs were collected with 2048 × 200 points and 8–64 scans for the different samples. One-dimensional 1H-NMR spectra were collected with 2048 points and 128 scans for all samples. For HSQC measurements, we used 0.2 mm uniformly labeled [15N]rPrP23–231 in 20 mm sodium phosphate buffer, pH 7.4, 100 mm NaCl, and a 10% D2O, 90% H2O mixture in the presence (at 1:1 molar ratio) or absence of SAF9343–59.
Transmission Electron Microscopy—Samples were adhered to a carbon-coated grid, blotted to remove excess material, and stained for 10 s with a 2% solution of uranyl acetate prepared in water. The images were digitally collected with a Zeiss EM 900 electron microscope (Carl Zeiss Inc.).
Neuroblastoma Cell (N2a) Culture and Cell Viability Assays—N2a cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 2% antibiotic (penicillin, erythromycin, and gentamicin) in a 5% CO2 atmosphere for 3 days and then transferred to a 96-well plate (∼ 5,000 cells/well). After transfer, the cells were exposed to samples containing rPrP23–231, N2aRNA, or rPrP-RNA complex for 3 days. An MTT assay was performed following the previously described protocol (30). MTT enters the cells by endocytosis and is reduced to formazan by NADH reductase and other enzymes in a reaction that can be measured spectro-photometrically. The amount of formazan reflects the reductive potential of the cytoplasm and the cell viability and generally shows good correlation with other viability tests (30, 39, 40). Live/dead assays were performed following the protocol provided by the kit (live/dead viability/cytotoxicity kit for mammalian cells; Invitrogen). Live cells were distinguished by the presence of ubiquitous intracellular esterase activity, which was determined by the enzymatic conversion of the virtually non-fluorescent cell-permeant calcein AM to the intensely fluorescent calcein. The polyanionic dye calcein is well retained within live cells, producing an intense uniform green fluorescence in live cells (ex/em =∼495 nm/∼515 nm). EthD-1 enters cells with damaged membranes and undergoes a 40-fold enhancement of fluorescence upon binding to nucleic acids, thereby producing a bright red fluorescence in dead cells (ex/em = ∼495 nm/∼635 nm). EthD-1 is excluded by the intact plasma membrane of live cells. All of the assays were done in triplicate. Live/dead images were generated in the Observer.Z1 Microscope (SN: 3834000373; Carl Zeiss Inc.).
Western Blotting—All of the protease-digested (+PK) samples were incubated with 20 μg/ml proteinase K (Sigma-Aldrich) for 1 h at 37 °C. SDS-PAGE was performed on 1.5-mm 15% polyacrylamide gels. The blocked membrane was incubated overnight at 4 °C with anti-PrP monoclonal antibody (SAF84; Cayman Chemical, MI) diluted 1:500. Following this incubation, the membrane was washed and incubated for 1 h at room temperature with secondary antibody, IgG-horseradish peroxidase-conjugated goat anti mouse (SC-2005; Santa Cruz Biotechnology, Santa Cruz, CA) diluted 1:200. The blot was stained with diaminobenzidine tetrahydrochloride and revealed with hydrogen peroxide (both from Sigma-Aldrich).
RESULTS
We first investigated the effects of RNA on the full-length recombinant PrP (rPrP23–231) in vitro. It has been shown that PrPC present in hamster brain homogenates interacts with RNA in vitro, acquiring protease resistance, and that RNase treatment can inhibit this conversion without changing the PrPC content (17). Questions remain unanswered, however, regarding the size and structure of the RNA that are necessary for interaction with PrPC and for catalysis of this conversion. In light of these results, we investigated the interaction of prokaryotic and eukaryotic RNA extracts and of two synthetic RNAs (SAF9343–59 and 18-mer opRNA) (Fig. 1 and supplemental Table S1) with rPrP23–231 and with two N-terminal rPrP deletion mutants (rPrPΔ51–90 and rPrPΔ32–121). The SAF9343–59 sequence was chosen based on previous work (22) that described the isolation of 2′-fluoropyrimidine-substituted RNA aptamers that bind selectively to disease-associated, β-sheet-rich isoforms of PrP. The SAF93 sequence was identified by Sayer et al. (23) as a high affinity PrP ligand; therefore, we selected the main binding domain (nucleotides 43–59) to perform our studies. The opRNA random sequence was selected because it has been suggested that PrP-nucleic acid interaction is guided by NA structure and not by its specific nucleotide sequence (22, 23). The local secondary structures of the synthetic RNAs used in this work were predicted with the program GeneBee, which indicated that both sequences adopt a hairpin-like structure (not shown). Thus we report the PrP binding behavior of two different base sequences with similar secondary structures.
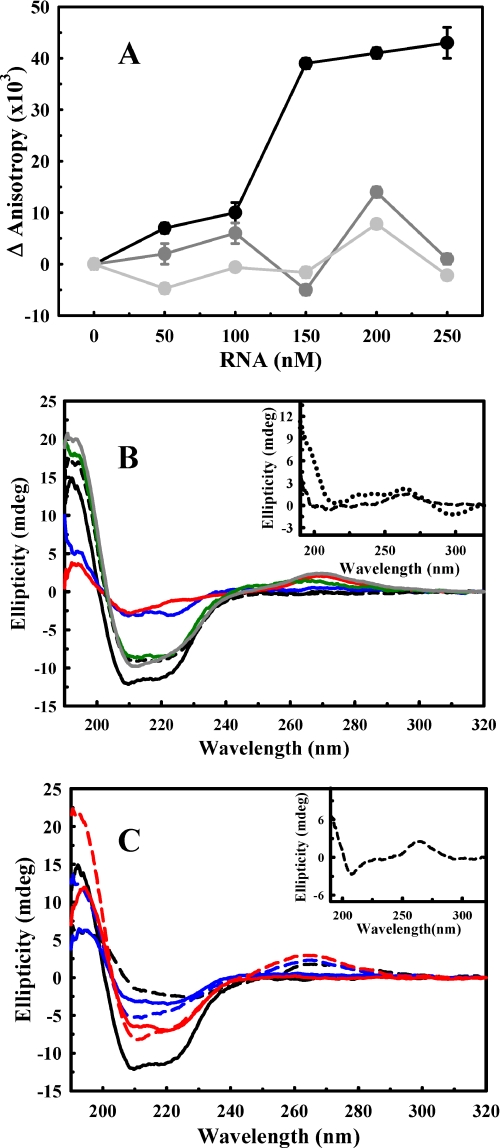
FIGURE 1.
RNA binding to rPrP and its effects on rPrP secondary structure. A, opRNA was titrated into 50 nm FITC-labeled rPrP23–231 (black), rPrPΔ51–90 (dark gray), or rPrPΔ32–121 (light gray). Anisotropy values were collected through a 3–69 filter (excitation was set at 490 nm). B, CD spectra of free rPrP23–231 (solid black line) and in the presence of opRNA (solid blue line) or SAF9343–59 (solid red line) and free rPrPΔ51–90 (dashed black line) and in the presence of opRNA (dark green line) or SAF9343–59 (dark gray line). The inset represents the CD spectra of opRNA (dashed black line) and SAF9343–59 (dotted black line) free in solution. C, CD spectra of free rPrP23–231 (solid black line) and in the presence of N2aRNA at 0.69 mg/ml (dashed black line); free rPrPΔ51–90 at 0.57 mg/ml (solid red line); rPrPΔ51–90 in the presence of N2aRNA at 0.57 mg/ml (dashed red line); rPrPΔ32–121 at 0.42 mg/ml (solid blue line) and rPrPΔ32–121 in the presence of N2aRNA at 0.42 mg/ml (dashed blue line). The inset represents the CD spectrum of free N2aRNA at 0.69 mg/ml. For B and C, protein and synthetic RNAs concentrations were 30 μm.
RNA Incubation with rPrP23–231 Leads to Protein Aggregation—We verified that incubation of the RNA extracts with recombinant PrP led to an increase in light scattering values for full-length rPrP, indicating that the protein aggregates upon RNA addition (Table 1 and supplemental Table S1). This result was obtained for incubation with both prokaryotic (from E. coli) and eukaryotic (from mammals, N2aRNA and vRNA; from mosquito cells, C6RNA; and from yeast, ScRNA) RNA extracts (supplemental Table S1). Interestingly, incubation of the rPrPΔ51–90 N-terminal deletion mutant with all of the RNAs investigated did not alter the LS values, revealing an important role for the PrP N-terminal region in the RNA interaction. We further selected the neuroblastoma cell-derived RNA (N2aRNA) to continue our studies because this RNA originates from the same tissue as our prion protein constructs (from mouse) (Figs. 1C and 2).
TABLE 1.
Summary of N2aRNA effects on rPrP23–231 and rPrPΔ51–90
|
Spectroscopic measurements
|
rPrP23–231
|
RNA digestion by RNase A
|
rPrPΔ51–90
|
|||
|---|---|---|---|---|---|---|
| –RNA | +RNA | Before rPrP incubation with RNA | After rPrP incubation with RNA | –RNA | +RNA | |
| ε 222 nma | –12.32 | –1.23 | – | – | –6.85 | –6.63 |
| LS/LS0b | 1 | 21.18 | 2.09 | 5.96 | 1 | 0.86 |
| FI/FI0b | 1 | 1.03 | 1.00 | 0.82 | 1 | 0.88 |
| ΔCM (cm–1)c | 0 | 227 | –35 | 90 | 0 | 6 |
Raw ellipticity values at 222 nm obtained from CD measurements
Normalized values obtained by dividing each recorded value (in the presence of N2aRNA) by the initial value (LS0, initial light scattering; FI0, initial fluorescence intensity) for free protein in solution
Changes in center of spectral mass values from the tryptophan emission spectra. N2aRNA digestion with RNase A was performed at room temperature for 1 h at pH 8.0 with a N2aRNa:RNase ratio of 100:1
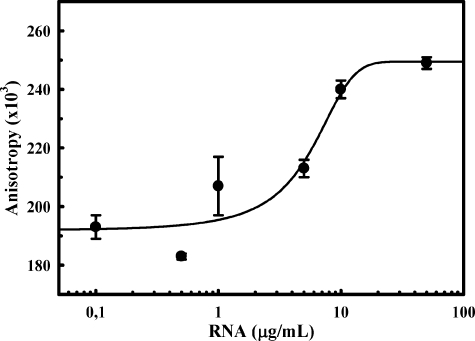
FIGURE 2.
N2aRNA binds rPrP23–231 with high affinity. N2aRNA was titrated into a 1.15 μg/ml (50 nm) FITC-rPrP23–231 solution, and the anisotropy values were collected through a 3–69 filter (excitation was set at 490 nm).
The rPrP N-terminal Region Is Necessary for RNA-induced Aggregation—We monitored the interaction of FITC-labeled rPrP23–231, rPrPΔ51–90, and rPrPΔ32–121 with opRNA by fluorescence anisotropy to further investigate the role of the N-terminal region in RNA binding to rPrP. Increasing concentrations of opRNA led to an increase in the anisotropy values only for RNA titration in rPrP23–231 solution, confirming the high affinity interaction between full-length PrP and RNA (Fig. 1A, black symbols). Binding of rPrP23–231 to N2aRNA was also observed, with a half-maximal effect at 5.95 μg/ml (Fig. 2). Consistent with measurements taken when rPrPΔ51–90 is incubated with different RNA extracts (supplemental Table S1), anisotropy data confirm the absence of interaction between the N-terminal deletion rPrP mutants and RNA in the investigated concentration range (Fig. 1A, gray symbols).
PrP-RNA Interaction Leads to Secondary and Tertiary Structural Changes Only for rPrP23–231—The changes in secondary structure of the prion protein constructs upon interaction with the different RNAs were analyzed by CD. To determine whether free RNA interferes with the CD spectra, far-UV CD spectra were collected for all RNA samples used. They all showed one negative peak (∼208 nm) and one positive peak (∼265 nm) (Fig. 1, B and C, insets). These results show a significant secondary structure signal from the nucleic acids that were investigated. Because the RNA structure may change upon rPrP binding, the RNA spectra were not subtracted from the spectra of the complex (PrP-RNA). Instead, the spectrum of the PrP-RNA complex is displayed together with the sum of each RNA spectrum and that of rPrP alone (supplemental Fig. S1). In this way, it is possible to infer whether the changes seen are due to modifications in the secondary structure of PrP or due only to the addition of the free RNA and rPrP spectra.
The addition of synthetic RNA oligonucleotides SAF9343–59 and opRNA (Fig. 1B) and of N2aRNA (Fig. 1C) led to immediate loss of secondary structure in rPrP23–231. Interestingly, changes in secondary structure caused by the synthetic RNA sequences were different from the effects observed for N2aRNA. Both SAF9343–59 and opRNA induced an increase in LS (supplemental Table S1), but this increase was much less marked than the one induced by N2aRNA (Table 1 and supplemental Table S1). This result indicates that, for the synthetic RNA sequences, an oligomeric species of limited size is populated. Unlike N2aRNA, in the presence of SAF9343–59 and opRNA, rPrP23–231 fluorescence emission intensity was reduced and shifts of the spectra (increase in center of spectral mass values) to more energetic wavelengths were also observed (supplemental Table S1). Because the N2aRNA is a pool of several RNAs with distinct sequences, sizes, and structures, differences between synthetic RNAs and the RNA pool are to be expected.
We observed that N2aRNA addition led to visible protein aggregation and that the CD spectrum of the complex showed immediate loss of α-helical content (at 222 nm) in the presence of N2aRNA (Fig. 1C and supplemental Fig. S1). This process is irreversible, because no significant gain in the secondary structure content was observed even after 7 days of incubation (not shown).
The aggregation of rPrP23–231 was confirmed by the increase in LS upon RNA addition (Table 1 and supplemental Table S1). The fluorescence emission spectrum was displaced toward more energetic wavelengths, as seen from the increase in CM values after N2aRNA incubation with rPrP23–231 (Table 1 and supplemental Table S1) but not when this RNA extract was incubated with rPrPΔ51–90 (Table 1). This result indicates protection of the tryptophan residues, which are mainly exposed to the solvent when rPrP23–231 is free in solution (35). Total fluorescence intensity did not change significantly in the presence of N2aRNA (Table 1 and supplemental Table S1).
There were no changes in secondary structure (α-helical content) in either of the rPrP N-terminal deletion mutants after incubation with opRNA, SAF9343–59, or N2aRNA, as shown in Fig. 1 (B and C) and supplemental Fig. S1 for rPrPΔ51–90 (not shown for rPrPΔ32–121). Furthermore, neither LS nor fluorescence emission changed significantly in either case upon RNA addition, indicating no changes in tertiary structure (Table 1, last column, and supplemental Table S1 for rPrPΔ51–90). These data agree with prior observations that suggest the importance of the N-terminal region to PrP-NA interactions (24, 25, 41, 42).
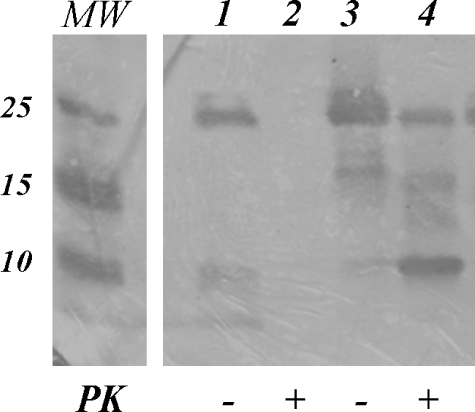
Treatment of rPrP23–231-N2aRNA Complex with Proteinase K—PrPC and PrPSc differ substantially in their conformations. Unlike PrPC, PrPSc is a multimeric assembly characterized by an increase in the amount of β-sheet structure and enhanced resistance to proteinase K (PK) digestion (3, 4). After observing rPrP23–231 changes in secondary structure and aggregation upon N2aRNA binding, we treated the aggregates with PK for 1 h at 37 °C (ratio PrP:PK = 100:1). The Western blotting shown in Fig. 3 revealed that PK treatment completely digested free rPrP23–231, but the rPrP23–231-N2aRNA complex was partially resistant to digestion. The same result was observed when PK resistance of the rPrP23–121-N2aRNA complex was monitored by SDS-PAGE and stained with silver (supplemental Fig. S2). Larger aggregates are also formed upon incubation with N2aRNA and present high resistance to PK digestion (supplemental Fig. S2B, asterisk). Treatment of other rPrP23–231-RNA complexes formed by other RNA extracts and synthetic sequences SAF9343–59 and opRNA, however, resulted in total digestion by PK (not shown). We performed the same assay for rPrPΔ51–90 and verified that this rPrP construction does not acquire protease resistance upon incubation with N2aRNA (supplemental Fig. S3), further indicating that the N-terminal mutants do not interact with the RNAs investigated here.
FIGURE 3.
rPrP23–231-N2aRNA resistance to PK digestion. Western blotting for PrP C-terminal domain. The samples were treated with PK at 37 °C for 1 h (ratio PrP:PK = 100:1), except where indicated (-). Lanes 1 and 2, rPrP23–231 at 5 μm; Lanes 3 and 4, rPrP23–231 at 5 μm + N2aRNA at 0.057 mg/ml. Molecular mass markers (MW) are shown on the left.
Treatment of N2aRNA with RNase Blocks the PrP-RNA Interaction Effects—Prior treatment of N2aRNA with RNase A abolished the effect of the nucleic acid on rPrP23–231 aggregation, as shown in Table 1. LS and fluorescence emission also returned to values close to that of the soluble, nonaggregated rPrP23–231. This indicates that the effects seen are specific for RNA interaction and not caused by interaction with free ribonucleotides in solution. In contrast, treatment with RNase A after RNA incubation with rPrP23–231 did not completely reverse aggregation and tertiary structure changes (Table 1), suggesting either that the rPrP aggregate formed can no longer be solubilized after RNA removal or that the RNA bound to PrP is protected from cleavage. To confirm the latter assumption we applied RNase-treated rPrP23–231-N2aRNA samples on agarose gel in the presence of ethidium bromide (supplemental Fig. S4). It is possible to visualize that larger aggregates are formed, and even after RNase treatment there is still RNA associated to the protein (supplemental Fig. S4, lane 4).
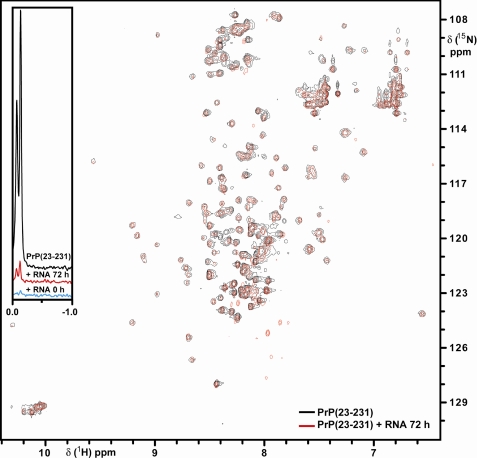
NMR Spectroscopy of PrP(23–231) and the Effect of RNA Addition—In Fig. 4, we present the NMR data obtained for rPrP23–231 in the presence or absence of RNA (SAF9343–59). We selected this RNA sequence because it is homogeneous and can be synthesized in sufficient quantities to perform the NMR measurements. The N2aRNA does not fulfill these characteristics. A small region of one-dimensional 1H NMR spectra taken for free rPrP23–231, rPrP23–231 immediately after RNA addition, and 3 days after RNA addition is shown in the inset of Fig. 4. These spectra were recorded and processed with the same spectral parameters so that the methyl signal shown could serve as a relative indication of soluble protein concentration.
FIGURE 4.
Characterization of rPrP23–231 and SAF9343–59 interaction by NMR spectroscopy. Superposition of the two-dimensional [15N,1H]HSQC spectra of rPrP(23–231) (black contours) and rPrP23–231 72 h after the addition of SAF9343–59 (red contours). The inset presents a region of one-dimensional 1H-NMR spectra of rPrP23–231 (black line), rPrP23–231 immediately after SAF9343–59 addition (blue line), and 72 h after RNA addition (red line).
According to the protein sequence, 288 HN cross-peaks are expected in the two dimensional [15N,1H]HSQC for rPrP23–231. We found 225 HN cross-peaks in the [15N,1H]HSQC of the sample prior to RNA supplementation. Immediately after RNA addition, there were no signals above the noise because of protein precipitation. It is worth mentioning that the precipitation can be visually detected during the addition of RNA. After a few days, the precipitation is no longer visible, and 3 days after RNA addition the sample presents 211 HN cross-peaks. As we noticed from the one-dimensional spectra, the amount of signal of this sample accounts for 8% of the signal of the initial sample without RNA. The spectra of PrP23–231 and PrP23–231 plus RNA are similar in regard to the chemical shift of several cross-peaks, indicating that both samples present the same three-dimensional structure. There are, however, many evident chemical shift differences between the spectra of these samples, in particular within the range of 7.8–8.6 ppm of 1H chemical shift. Furthermore, the area within 7.6–8.4 ppm of 1H and 124–129 ppm of 15N chemical shift has many additional cross-peaks. This 1H spectral range is populated by many amide cross-peaks of unfolded polypeptide backbone.
Taken together, these NMR data indicate that immediately after the addition of RNA, rPrP23–231 undergoes severe aggregation, which turns slowly into a soluble form of rPrP23–231. This is a new form of rPrP, which closely resembles the globular region of the free protein but with some modification in the unfolded N-terminal region of rPrP23–231. It is worth mentioning that the N terminus (residues 23 to ∼120) accounts for most of the PrP flexibly disordered behavior, whereas the C terminus (residues 121–231) encompasses a globular fold (35).
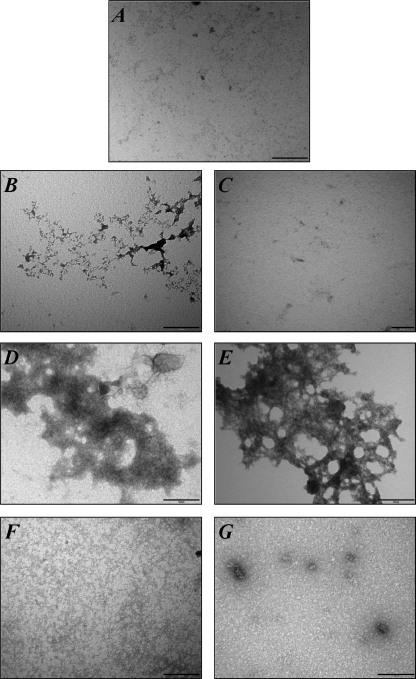
Ultrastructural Analysis of rPrP-RNA Complexes—To characterize possible morphological differences between the aggregates obtained after incubation with the RNA extract, N2aRNA, and with the small oligonucleotide sequence, SAF9343–59, we collected transmission electron microscopy images of the samples. Recombinant PrP23–231 was incubated with the aforementioned RNAs, and the transmission electron microscopy grids were prepared immediately. Fig. 5 displays both complexes, and we observed that rPrP forms amorphous aggregates upon incubation with RNA that are not present in soluble, RNA-free rPrP samples. Both aggregates appear very similar, but the one generated by the SAF sequence seems to adopt a more organized structure. These slight differences may account for the different structural changes elicited by the RNAs investigated.
FIGURE 5.
Electron microscopy of rPrP23–231 and rPrPΔ51–90 in the presence of RNA. A, rPrP23–231 at 2 μm (46 μg/ml). B, N2aRNA at 23 μg/ml. C, SAF9343–59 2 μm. D, rPrP23–231 at 2 μm (46 μg/ml) in the presence of N2aRNA at 23 μg/ml. E, rPrP23–231 at 2 μm in the presence of SAF9343–59 at 2 μm. F, rPrPD51–90 at 2 μm (38 μg/ml). G, rPrPΔ51–90 at 2 μm (38 μg/ml) in the presence of N2aRNA at 19 μg/ml. Scale bars, 0.2 μm.
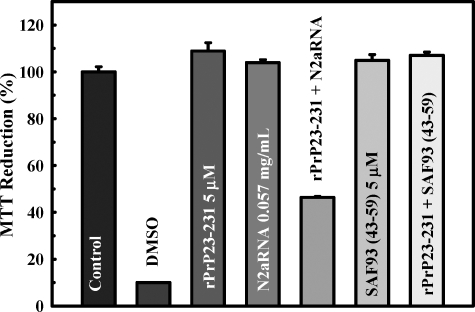
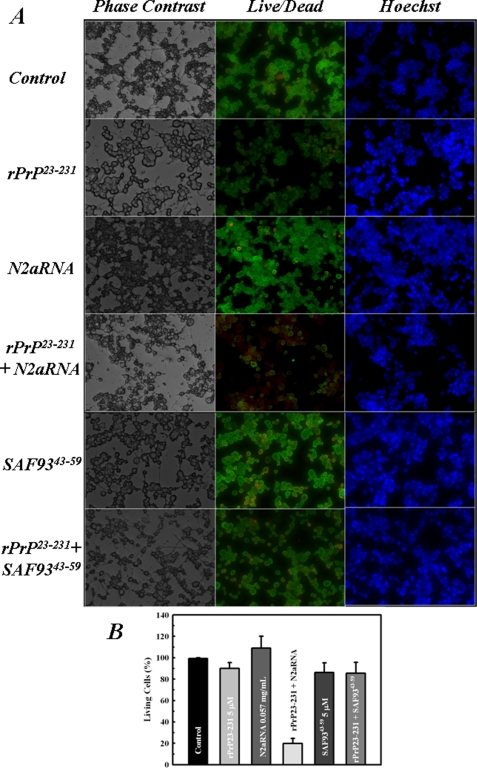
PrP-RNA Complex Alters Cultured Cell Viability—A series of aggregates, oligomers, and fibers generated in vitro have been shown to be toxic to cultured cells, leading to death by apoptosis (32), but there is no report of toxic PrPSc/res generated in vitro using total recombinant PrP purified from a heterologous system. After structural characterization of the rPrP23–231-RNA interaction, we investigated whether the aggregates formed upon N2aRNA or SAF9343–59 interaction with rPrP23–231 were toxic to cultured N2a cells using MTT reduction (30) and viability assays to measure cellular survival. As shown in Fig. 6, only the rPrP23–231-N2aRNA complex was able to alter the MTT reduction pattern in cultured neuroblastoma cells, leading to a ∼60% decrease in MTT reduction compared with rPrP23–231 alone. The complex formed with the synthetic sequence SAF9343–59 did not affect the cultures (Fig. 6), and neither did the samples containing only RNA molecules, which suggests that the PrP changes induced by RNA cause formation of a toxic PrP species. MTT reduction results were corroborated by viability assays in the presence of SAF9343–59, opRNA, and N2aRNA. The rPrP-N2aRNA complex was the most toxic to cells in culture (Fig. 7), and the rPrP complexes with SAF9343–59 (Fig. 7) or opRNA did not show significant toxic effects. The free opRNA sequence was already toxic to cells, but the addition of rPrP23–231 did not increase the observed cytotoxicity (not shown).
FIGURE 6.
rPrP23–231-N2aRNA complex alters MTT reduction in cultured neuroblastoma cells. Samples containing rPrP23–231, RNA molecules, or protein-RNA complexes were added to the culture medium 3 days before MTT addition. MTT reduction assay was performed as previously described (29). The following concentrations were used: rPrP23–231 at 0.115 mg/ml (5 μm), N2aRNA at 0.057 mg/ml, rPrP-N2aRNA (0.115 mg/ml:0.057 mg/ml) complex, SAF9343–59 at 5 μm, and rPrP23–231:SAF9343–59 (5 μm:5 μm) complex. For simplicity, the results are expressed as percentages of living cells relative to the value for control cells (cells in culture medium only). This expression is 100 × (ODsample × ODblank)/(ODcontrol × ODblank). The ODblank was established from the average of the wells containing only Dulbecco's modified Eagle's medium with no cells. ODcontrol was established from the average of wells containing nontreated cells. DMSO, dimethyl sulfoxide.
FIGURE 7.
Only rPrP23–231-N2aRNA complex is toxic to cultured neuroblastoma cells. A, cell viability measured by live/dead assay. The following concentrations were used: rPrP23–231 at 0.115 mg/ml (5 μm), N2aRNA at 0.057 mg/ml, rPrP-N2aRNA (0.115 mg/ml:0.057 mg/ml) complex, SAF9343–59 at 5 μm, and rPrP23–231:SAF9343–59 (5 μm:5 μm) complex. B, statistics of viability assay presented in A. For simplicity, the results are expressed as the percentages of living cells relative to the value for control cells (cells in culture medium only).
All of these results show that prion protein interacts with RNA molecules and that this interaction leads to protein aggregation. There is still no single sequence identified that would specifically bind to rPrP, but we have shown that the N-terminal prion protein domain is responsible for RNA recognition and binding. For DNA binding, both the globular and N-terminal domains are important (15).
DISCUSSION
Although the protein-only hypothesis is currently the most accepted explanation for the development of transmissible spongiform encephalopathies (1), the evidence for an additional cofactor in the PrPC conversion is accumulating (7–10, 17, 42). The physiological role of prion protein interaction with nucleic acids has attracted special attention in the last five years, with particular emphasis on the potential for new therapeutic strategies (27, 43). We have shown that recombinant murine prion protein binds to nucleic acid sequences and acquires higher β-sheet content but remains soluble, assuming a scrapie-like form (8, 44). More recently, we have mapped the regions that are directly involved in DNA binding by PrP (15), which revealed that both the globular and N-terminal domains participate in the binding event. This result clarified the importance of the PrP globular domain for DNA recognition, but it also showed for the first time that the N-terminal domain is involved in stabilization of the complex. Since then, new information about the possible physiological relevance of prion-nucleic acid complexes in vivo has appeared (13, 17, 19, 45). The use of NA aptamers for the diagnosis of prion diseases is another interesting approach that takes advantage of the selective PrP-NA interaction. It has been reported that some aptamers can interact with specific isoforms of the prion protein, recognizing either the cellular or the scrapie form of PrP in vitro (20, 23, 46). It remains a challenge to develop such an assay for PrPSc identification in vivo.
Here we show that RNA sequences can also interact with the prion protein with high affinity but with different specificities and that they lead to different changes in the secondary and tertiary structure of recombinant PrP compared with DNA binding. The interaction of prion protein with ribonucleic acid has been investigated by other groups; it has been shown that PrP can acquire protease resistance upon RNA binding (17) and that scrapie and cellular PrP isoforms can bind with different affinities to some highly structured RNA sequences (19–21). This property has allowed specific RNA aptamers to be constructed to aid in the early diagnosis of prion diseases in humans and animals. Although much effort has been devoted to identifying a key sequence or effect of RNA interaction with the prion protein, this subject remains controversial. First, there is no single, specific sequence that binds to PrP, nor is there a single nucleic acid-binding domain in this protein. Moreover, the minimum number of bases required for efficient binding of PrP by RNA is unknown. Therefore, the aim of the present work was to investigate in more detail what happens to the prion protein structure during interaction with RNA and also to compare the observed changes with previous data on the PrP-DNA interaction (8, 15). We have seen that these interactions, although identical in some respects, display substantial differences.
We have shown that PrP interacts with RNA with high affinity (nanomolar range) and that this interaction leads to an insoluble aggregated protein. It is noteworthy that PrP is less prone to aggregation when it interacts with double-stranded DNAs (8, 15). Moreover, the prion protein (N-terminal) deletion mutants do not seem to interact with any of the RNA sequences investigated. RNA-induced aggregation of prion protein depends on the N-terminal region of this protein, indicating that at least part of the 51–90 PrP region is responsible for this process. It has been reported that truncation of the PrP N-terminal region affects formation of protease-resistant PrP (PrP-res) in vitro (47). Reduced formation of PrP-res with deletion of N-terminal residues relative to full-length hamster PrP (47) may be related to RNA binding ability in vivo because N-terminal deletion mutants do not bind RNA and are also not toxic to cultured cells. This differs from what was observed with the PrP-DNA interaction (15), suggesting that the role of these nucleic acids in prion biology must be different. In an early publication, Gabus et al. (26) found that the human prion protein has DNA strand transfer properties comparable with the retroviral nucleocapsid protein and that it forms large nucleoprotein complexes upon binding to DNA. The same group found that the huPrP(23–144) fragment could bind DNA and have strand transfer activity (26). Our previous NMR study demonstrated that most of the changes in chemical shift upon DNA binding were in the N terminus domain and in helix 1 (15).
Interestingly, the NMR HSQC measurements revealed that, although there is immediate aggregation of rPrP23–231 upon RNA addition, a few days after incubation with RNA, the rPrP recovers most of its original fold (Fig. 4). This result indicates that the changes induced by this synthetic RNA are partially reversible, but most importantly, the appearance of additional cross-peaks in the HSQC spectrum indicates that part of the protein N-terminal domain is modified after interaction with RNA (Fig. 4). The NMR data lead us to conclude that, although the overall fold of rPrP is recovered ∼3 days after RNA addition, significant structural changes are now present and can reveal important features about the PrP interaction with ribonucleic acids.
We previously reported that α-helical rPrP is more hydrated and has a larger solvent-accessible surface area than aggregated β-rPrP as measured by pressure perturbation calorimetry (48), and it may be that nucleic acid binding to cellular rPrP leads to similar dehydration effects. These changes in structure of the prion protein when complexed to the nucleic acid would be typical of nucleic acid chaperones (10, 41, 42, 49).
To investigate whether the aggregates formed upon RNA binding are toxic to N2a cells in culture, we performed cell viability assays. Interestingly, the aggregates formed upon PrP incubation with total RNA extract from N2a cells were significantly cytotoxic, leading to an approximately 60% loss in cell viability (Fig. 6). In contrast, we did not observe significant toxicity when recombinant PrP aggregates were induced by small synthetic oligonucleotides (opRNA or SAF).
Our findings obtained when PrP protein binds either to DNA or RNA show that the heterogeneous mixture of RNA extracted from neuroblastoma cells was the only one to trigger toxicity and massive aggregation. The small RNAs were able to bind PrP, also leading to protein aggregation (Fig. 5, B and D), but the oligomeric species formed presented no toxicity (Figs. 6 and 7).
The component of the N2A RNA extract that leads to aggregation and further toxicity remains to be identified, but we suppose that a specific RNA sequence or structure present in this preparation may be responsible for this effect. The local secondary structures of the synthetic RNAs, based on the prediction program GeneBee, were predicted to be hairpins. Although the SAF sequence has more guanine (∼39% versus ∼22% for opRNA), they are otherwise similar in base content. The guanine base is expected to contribute to a greater number of hydrogen bonds with the binding protein (50), but both RNA sequences displayed a very similar pattern of interaction with the prion protein, and this base composition difference would also not explain the toxicity observed upon incubation with N2aRNA. Moreover, because the N2aRNA pool is very heterogeneous, it is still not possible to infer whether there it contains a specific RNA secondary structure responsible for rPrP binding and conversion to a toxic species. Altogether, our results reveal that there is selectivity in the species generated by interaction with different molecules of RNA. Because the catalytic effect of RNA on the PrPC → PrPSc conversion may depend on the RNA sequence, small RNA molecules may exert a protective effect.
Supplementary Material
Acknowledgments
We thank Prof. Martha M. Sorenson for carefully reading the manuscript, Dr. Vilma R. Martins for constructing and providing the rPrPs expression vectors, Aline Brando Lima for the help with western blotting technique, and Emerson Gonçalves for excellent technical assistance.
This work is dedicated to Leopoldo de Meis in commemoration of his 70th birthday.
This work was supported by grants from the Conselho Nacional de Desenvolvimento Científico e Tecnológico, the Millenium Institute for Structural Biology in Biomedicine and Biotechnology (Conselho Nacional de Desenvolvimento Científico e Tecnológico Millenium Program), Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (to J. L. S. and Y. C.), Financiadora de Estudos e Projetos of Brazil, an international grant from the International Centre for Genetic Engineering and Biotechnology (to J. L. S.), and a grant from L′Oréal, and Fundação Universitária José Bonifácio grant 13145-8 (to Y. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Author's Choice—Final version full access.
The on-line version of this article (available at http://www.jbc.org) contains supplemental data, supplemental Table S1, and supplemental Figs. S1–S4.
Footnotes
The abbreviations used are: PrP, prion protein; rPrP, recombinant prion protein; HSQC, heteronuclear single quantum coherence; LS, light scattering; N2aRNA, total RNA extracted from cultured neuroblastoma cells; NA, nucleic acid; FITC, fluorescein isothiocyanate; MTT, 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide; PK, proteinase K.
References
- 1.Prusiner, S. B. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 13363-13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguzzi, A., and Polymenidou, M. (2004) Cell 116 313-327 [DOI] [PubMed] [Google Scholar]
- 3.Caughey, B. W., Dong, A., Bhat, K. S., Ernst, D., Hayes, S. F., and Caughey, W. S. (1991) Biochemistry 30 7672-7680 [DOI] [PubMed] [Google Scholar]
- 4.Pan, K. M., Baldwin, M., Nguyen, J., Gasset, M., Serban, A., Groth, D., Mehlhorn, I., Huang, Z., Fletterick, R. J., Cohen, F. E., and Prusiner, S. B. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 10962-10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weissmann, C. (2004) Nat. Rev. Microbiol. 2 861-871 [DOI] [PubMed] [Google Scholar]
- 6.Bueler, H., Aguzzi, A., Sailer, A., Greiner, R. A., Autenried, P., Aguet, M., and Weissmann, C. (1993) Cell 73 1339-1347 [DOI] [PubMed] [Google Scholar]
- 7.Telling, G. C., Scott, M., Mastrianni, J., Gabizon, R., Torchia, M., Cohen, F. E., DeArmond, S. J., and Prusiner, S. B. (1995) Cell 83 79-90 [DOI] [PubMed] [Google Scholar]
- 8.Cordeiro, Y., Machado, F., Juliano, L., Juliano, M. A., Brentani, R. R., Foguel, D., and Silva, J. L. (2001) J. Biol. Chem. 276 49400-49409 [DOI] [PubMed] [Google Scholar]
- 9.Caughey, B., and Kocisko, D. A. (2003) Nature 425 673-674 [DOI] [PubMed] [Google Scholar]
- 10.Silva, J. L., Lima, L. M. T. R., Foguel, D., and Cordeiro, Y. (2008) Trends Biochem. Sci. 33 132-140 [DOI] [PubMed] [Google Scholar]
- 11.Baskakov, I. V., Legname, G., Prusiner, S. B., and Cohen, F. E. (2001) J. Biol. Chem. 276 19687-19690 [DOI] [PubMed] [Google Scholar]
- 12.Cordeiro, Y., and Silva, J. L. (2005) Protein Pept. Lett. 12 251-255 [DOI] [PubMed] [Google Scholar]
- 13.Caughey, B., Brown, K., Raymond, G. J., Katzenstein, G. E., and Thresher, W. (1994) J. Virol. 68 2135-2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horonchik, L., Tzaban, S., Ben-Zaken, O., Yedidia, Y., Rouvinski, A., Papy-Garcia, D., Barritault, D., Vlodavsky, I., and Taraboulos, A. (2005) J. Biol. Chem. 280 17062-17067 [DOI] [PubMed] [Google Scholar]
- 15.Lima, L. M., Cordeiro, Y., Tinoco, L. W., Marques, A. F., Oliveira, C. L., Sampath, S., Kodali, R., Choi, G., Foguel, D., Torriani, I., Caughey, B., and Silva, J. L. (2006) Biochemistry 45 9180-9187 [DOI] [PubMed] [Google Scholar]
- 16.Nandi, P. K., Leclerc, E., Nicole, J. C., and Takahashi, M. (2002) J. Mol. Biol. 322 153-161 [DOI] [PubMed] [Google Scholar]
- 17.Deleault, N. R., Lucassen, R. W., and Supattapone, S. (2003) Nature 425 717-720 [DOI] [PubMed] [Google Scholar]
- 18.Nandi, P. K., and Nicole, J. C. (2004) J. Mol. Biol. 344 827-837 [DOI] [PubMed] [Google Scholar]
- 19.Deleault, N. R., Georghegan, J. C., Nishina, K., Kascsak, R., Williamson, R. A., and Supattapone, S. (2005) J. Biol. Chem. 280 26873-26879 [DOI] [PubMed] [Google Scholar]
- 20.Deleault, N. R., Harris, B. T., Rees, J. R., and Supattapone, S. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 9741-9746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adler, V., Zeiler, B., Kryukov, V., Kascsak, R., Rubenstein, R., and Grossman, A. (2003) J. Mol. Biol. 332 47-57 [DOI] [PubMed] [Google Scholar]
- 22.Rhie, A., Kirby, L., Sayer, N., Wellesley, R., Disterer, P., Sylvester, I., Gill, A., Hope, J., James, W., and Tahiri-Alaoui, A. (2003) J. Biol. Chem. 278 39697-39705 [DOI] [PubMed] [Google Scholar]
- 23.Sayer, N. M., Cubin, M., Rhie, A., Bullock, M., Tahiri-Alaoui, A., and James, W. (2004) J. Biol. Chem. 279 13102-13109 [DOI] [PubMed] [Google Scholar]
- 24.Weiss, S., Proske, D., Neumann, M., Groschup, M. H., Kretzschmar, H. A., Famulok, M., and Winnacker, E. L. (1997) J. Virol. 71 8790-8797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sekiya, S., Noda, K., Nishikawa, F., Yokoyama, T., Kumar, P. K., and Nishikawa, S. (2006) J. Biochem. (Tokyo) 139 383-390 [DOI] [PubMed] [Google Scholar]
- 26.Gabus, C., Auxilien, S., Péchoux, C., Dormont, D., Swietnicki, W., Morillas, M., Surewicz, W., Nandi, P., and Darlix, J. L. (2001) J. Mol. Biol. 307 1011-1021 [DOI] [PubMed] [Google Scholar]
- 27.King, D. J., Safar, J. G., Legname, G., and Prusiner, S. B. (2007) J. Mol. Biol. 369 1001-1014 [DOI] [PubMed] [Google Scholar]
- 28.Lambert, M. P., Barlow, A. K., Chromy, B. A., Edwards, C., Freed, R., Liosatos, M., Morgan, T. E., Rozovsky, I., Trommer, B., Viola, K. L., Wals, P., Zhang, C., Finch, C. E., Krafft, G. A., and Klein, W. L. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 6448-6453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gosavi, N., Lee, H. J., Lee, J. S., Patel, S., and Lee, S. J. (2002) J. Biol. Chem. 277 48984-48992 [DOI] [PubMed] [Google Scholar]
- 30.Reixach, N., Deechongkit, S., Jiang, X., Kelly, J. W., and Buxbaum, J. N. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 2817-2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castilla, J., Saa, P., Hetz, C., and Soto, C. (2005) Cell 121 195-206 [DOI] [PubMed] [Google Scholar]
- 32.Novitskaya, V., Bocharova, O. V., Bronstein, I., and Baskakov, I. V. (2006) J. Biol. Chem. 281 13828-13836 [DOI] [PubMed] [Google Scholar]
- 33.Zou, W. Q., and Gambetti, P. (2005) Cell 121 155-157 [DOI] [PubMed] [Google Scholar]
- 34.Cordeiro, Y., Kraineva, J., Gomes, M. P. B., Lopes, M. H., Martins, V. R., Lima, L. M. T. R., Foguel, D., Winter, R., and Silva, J. L. (2005) Biophys. J. 89 2667-2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riek, R., Hornemann, S., Wider, G., Glockshuber, R., and Wuthrich, K. (1997) FEBS Lett. 413 282-288 [DOI] [PubMed] [Google Scholar]
- 36.Windl, O., Lorenz, H., Behrens, C., Romer, A., and Kretzschmar, H. A. (1999) J. Gen. Virol. 80 15-21 [DOI] [PubMed] [Google Scholar]
- 37.Lim, F., Downey, T. P., and Peabody, D. S. (2001) J. Biol. Chem. 276 22507-22513 [DOI] [PubMed] [Google Scholar]
- 38.Keller, R. (2005) Optimizing the Process of Nuclear Magnetic Resonance Spectrum Analysis and Computer-Aided Resonance Assignment Diss. ETH No. 15947, Swiss Federal Institute of Technology, Zurich
- 39.Liu, Y., Peterson, D. A., Kimura, H., and Schubert, D. (1997) J. Neurochem. 69 581-593 [DOI] [PubMed] [Google Scholar]
- 40.Loske, C., Neumann, A., Cunningham, A. M., Nichol, K., Schinzel, R., Riederer, P., and Munch, G. (1998) J. Neural Transm. 105 1005-1015 [DOI] [PubMed] [Google Scholar]
- 41.Gabus, C., Derrington, E., Leblanc, P., Chnaiderman, J., Dormont, D., Swietnicki, W., Morillas, M., Surewicz, W. K., Marc, D., Nandi, P., and Darlix, J. L. (2001) J. Biol. Chem. 276 19301-19309 [DOI] [PubMed] [Google Scholar]
- 42.Supattapone, S. (2004) J. Mol. Med. 82 348-356 [DOI] [PubMed] [Google Scholar]
- 43.Kocisko, D. A., Vaillant, A., Lee, K. S., Arnold, K. M., Bertholet, N., Race, R. E., Olsen, E. A., Juteau, J. M., and Caughey, B. (2006) Antimicrob. Agents Chemother. 50 1034-1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cordeiro, Y., Lima, L. M., Gomes, M. P., Foguel, D., and Silva, J. L. (2004) J. Biol. Chem. 279 5346-5352 [DOI] [PubMed] [Google Scholar]
- 45.Zou, W. Q., Zheng, J., Gray, D. M., Gambetti, P., and Chen, S. G. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 1380-1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takemura, K., Wang, P., Vorberg, I., Surewicz, W., Priola, S. A., Kanthasamy, A., Pottathil, R., Chen, S. G., and Sreevatsan, S. (2006) Exp. Biol. Med. 231 204-214 [DOI] [PubMed] [Google Scholar]
- 47.Lawson, V. A., Priola, S. A., Meade-White, K., Lawson, M., and Chesebro, B. (2004) J. Biol. Chem. 279 13689-13695 [DOI] [PubMed] [Google Scholar]
- 48.Cordeiro, Y., Kraineva, J., Ravindra, R., Lima, L. M., Gomes, M. P., Foguel, D., Winter, R., and Silva, J. L. (2004) J. Biol. Chem. 279 32354-32359 [DOI] [PubMed] [Google Scholar]
- 49.Cristofari, G., and Darlix, J. L. (2002) Prog. Nucleic Acids Res. Mol. Biol. 72 223-268 [DOI] [PubMed] [Google Scholar]
- 50.Luscombe, N. M., Laskowski, R. A., and Thornton, J. M. (2001) Nucleic Acids Res. 29 2860-2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.