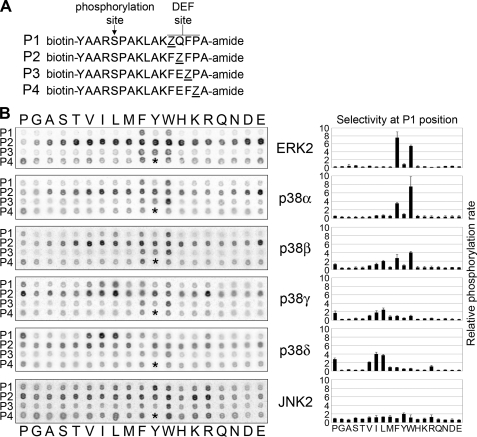
FIGURE 3.
Identification of MAPK DEF site motifs using a novel positional scanning peptide library. A, peptide library incorporates 19 amino acid substitutions (Z) at each of the four positions within the DEF site (P1–P4). B, peptides were subjected to phosphorylation by the MAPK in the presence of [γ-33P]ATP followed by capture on streptavidin membrane and exposure to a phosphor screen (left panel). p38 MAPK isoforms displayed unique selectivity at the P1 position. The p38α selectivity is similar to ERK2, with a preference for aromatic residues at P1, although p38δ favors aliphatic residues at P1. p38β and p38γ displayed intermediate P1 selectivity with p38β more similar to p38α and p38γ more similar to p38δ. JNK2 did not display a strong sequence preference in the DEF site (<2-fold difference for any substitution). The P4 Tyr peptide was not included because of failed synthesis (space indicated with an asterisk). Histograms depict the quantified radiolabel incorporation for the P1 position. The selectivity value is calculated as the level of phosphorylation of a given peptide divided by the average level of phosphorylation for all 19 peptides. Error bars represent the standard deviation from three separate experiments.