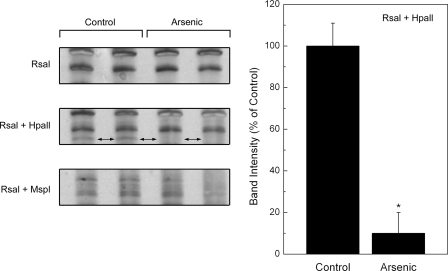
FIGURE 7.
Selected areas of DNA methylation in control or arsenic-exposed RWPE-1 cells at 4 weeks of exposure using arbitrarily primed PCR after restriction enzyme digestions. Shown are details of a representative gel with an area of distinct DNA hypomethylation in arsenic-treated DNA (left panels, two-headed arrows) and densitometric analysis (n = 3) of this band (right panel). See methods for details. An asterisk indicates a significant difference (p ≤ 0.05) from control. Unmethylated DNA is not protected from digestion by methylation-sensitive HpaII restriction enzyme resulting in a loss of DNA for PCR amplification. Digestion with RsaI + MspI would also result in no PCR amplification regardless of methylation status. Digestion with RsaI alone and RsaI + MspI were used as controls to determine whether there was differential methylation.