Figure 3.
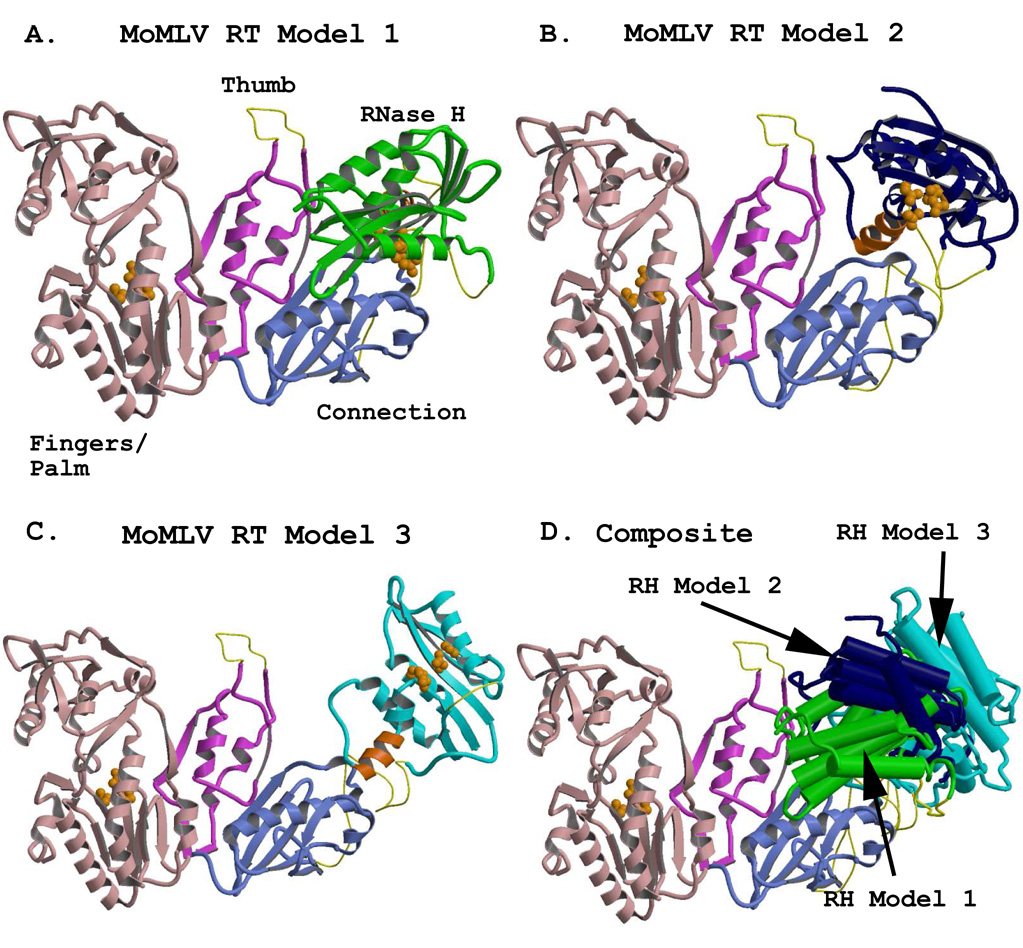
Representations of the full-length MoMLV RT structure, showing alternate positions of the RNase H domain. The models were generated using combinations of the crystal structure coordinates of PDB files 1RW3 and 2HB5. The fingers/palm, thumb, and connection subdomains are colored in light pink, fuscia, and light blue, respectively, and are rendered from the crystal structure coordinates of 1RW3. The RNase H domains were rendered from the coordinates of the crystal structure 2HB5. An added C-helix was modeled into the RNase H domains, and appears in orange. Regions missing from the reported crystal structures (see Table 3) are modeled with thin yellow connectors. Orange spheres represent the residues of the polymerase active site in the Fingers/Palm (F/P) and those of the DEDD active site in the RNases H. A) The full length MoMLV RT of Model 1 with its RNase H in green, occupying the position most closely fitted to the experimental electron density for the lRW3 crystal structure. B) MoMLV RT Model 2, with its RNase H in dark blue, rotated to a position +30° in y and +20° in x from its experimental locus, exposing both the C-helix and the RNase H active site. C) MoMLV RT Model 3, with its RNase H colored in cyan, superpositioned onto the placement of the RNase H as it occurs in the observed crystal structure of HIV-1 RT (PDB file 1HYS) and roated an extra 1.5° in the z direction. D) A superpositioning of the structures shown in A–C, with the structural elements of the RNases H rendered with cylinders for clarity. The colors of the RNases H correspond to those seen in the preceding panels.
