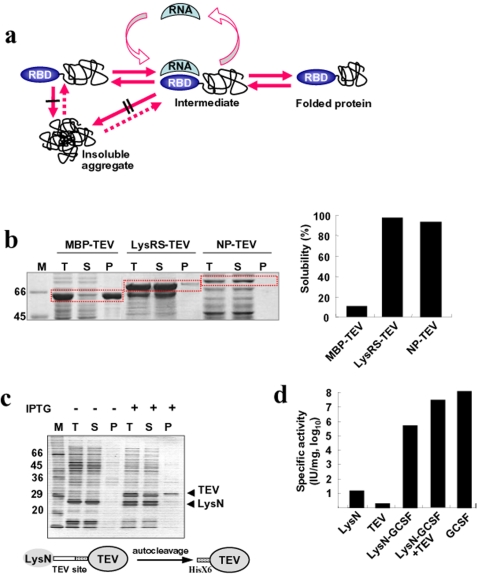
Figure 1. Development of RBPs as solubility enhancers.
(a) Proposed model for RNA binding-mediated protein folding. Both the folded RBD at N-terminal position and bound RNA prevent inter-molecular interactions among folding intermediates, leading to soluble expression and favoring kinetic network into productive folding. The number of black bars (| and ∥) represents the extent of aggregation inhibition. (b) The comparison of solubility-enhancing ability by RBP with that of MBP. E. coli lysyl tRNA synthetase (LysRS) and influenza virus nucleoprotein (NP) were used as RBP to monitor the soluble expression of tobacco etch virus (TEV) protease. The solubility-enhancing ability of RBP was compared to that of MBP. The fusion proteins were expressed at 37°C and their solubility was analyzed by SDS-PAGE. M, T, S, and P represent molecular weight marker, total lysates, soluble fraction, and insoluble fraction, respectively. (c) Autocatalytic cleavage of LysN-TEV containing TEV cleavage sequence between LysN and TEV protease in E. coli cytosol. Non-induced (−) and IPTG induced (+) cell extracts were analyzed by SDS-PAGE. The uncleaved LysN-TEV was not detected clearly on SDS-PAGE due to efficient cleavage. (d) Cell proliferation assay of GCSF expressed as LysN-GCSF. The purified TEV protease described in Figure 1c was used to cleave the purified LysN-GCSF. The purified LysN, TEV protease, and LysN-GCSF before and after cleavage with TEV protease were compared with the GCSF standard in the cell proliferation assay as described in Methods.